Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity
Abstract
:1. Introduction
2. Viral Glycoproteins—Essential Components for Viral Interactions with Host Cells
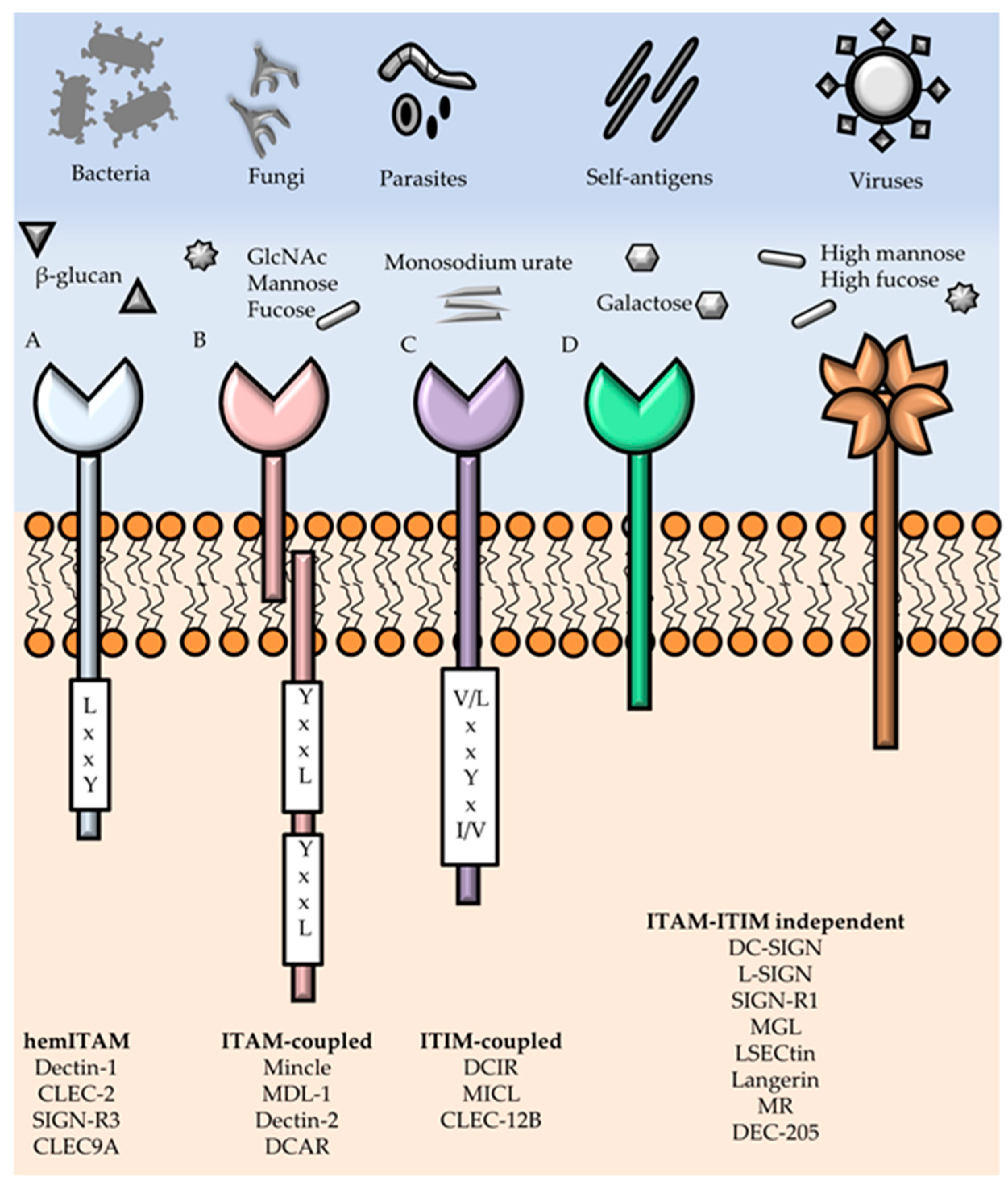
3. Myeloid C-type Lectins Receptors—Pattern Recognition Receptors in Innate Immunity
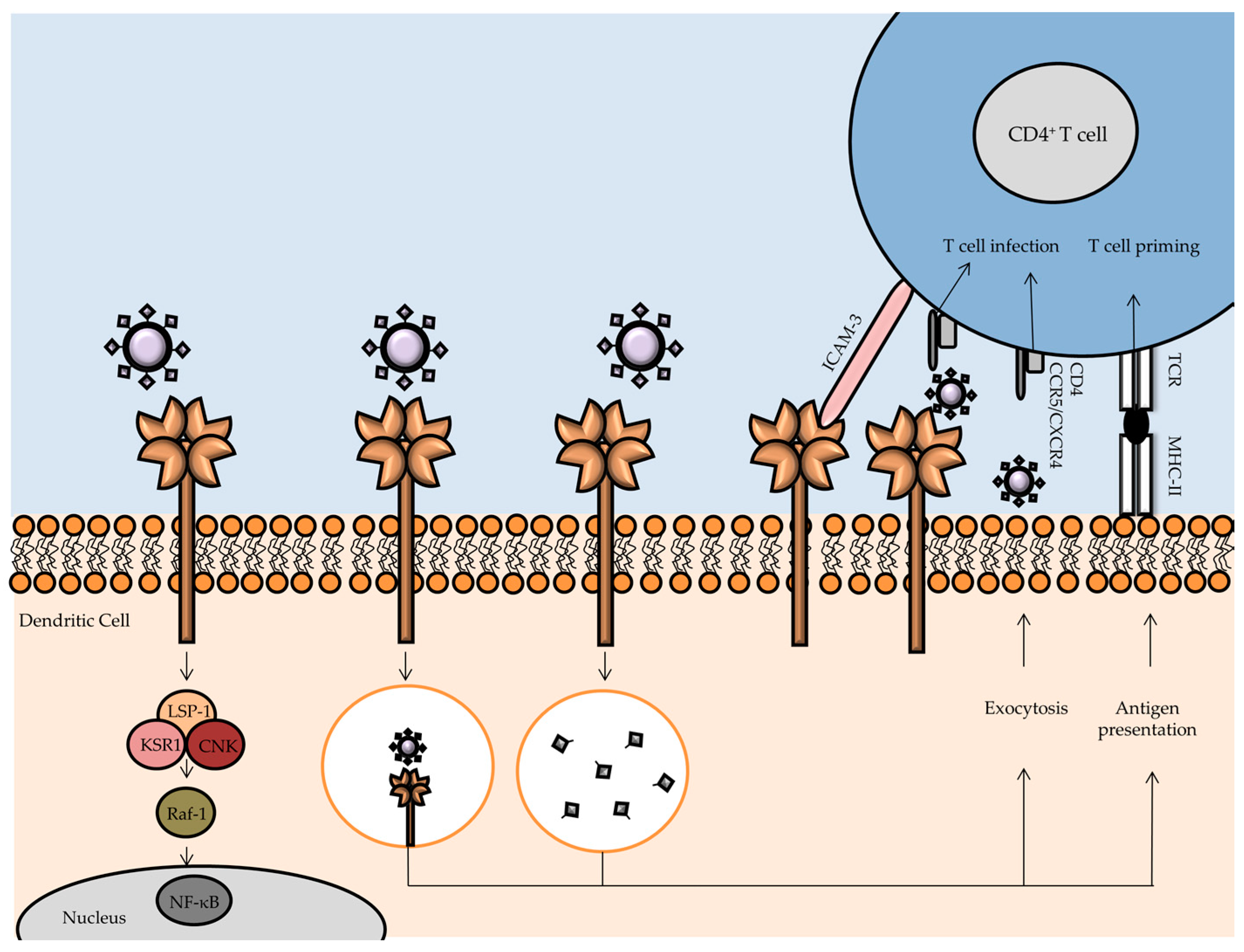
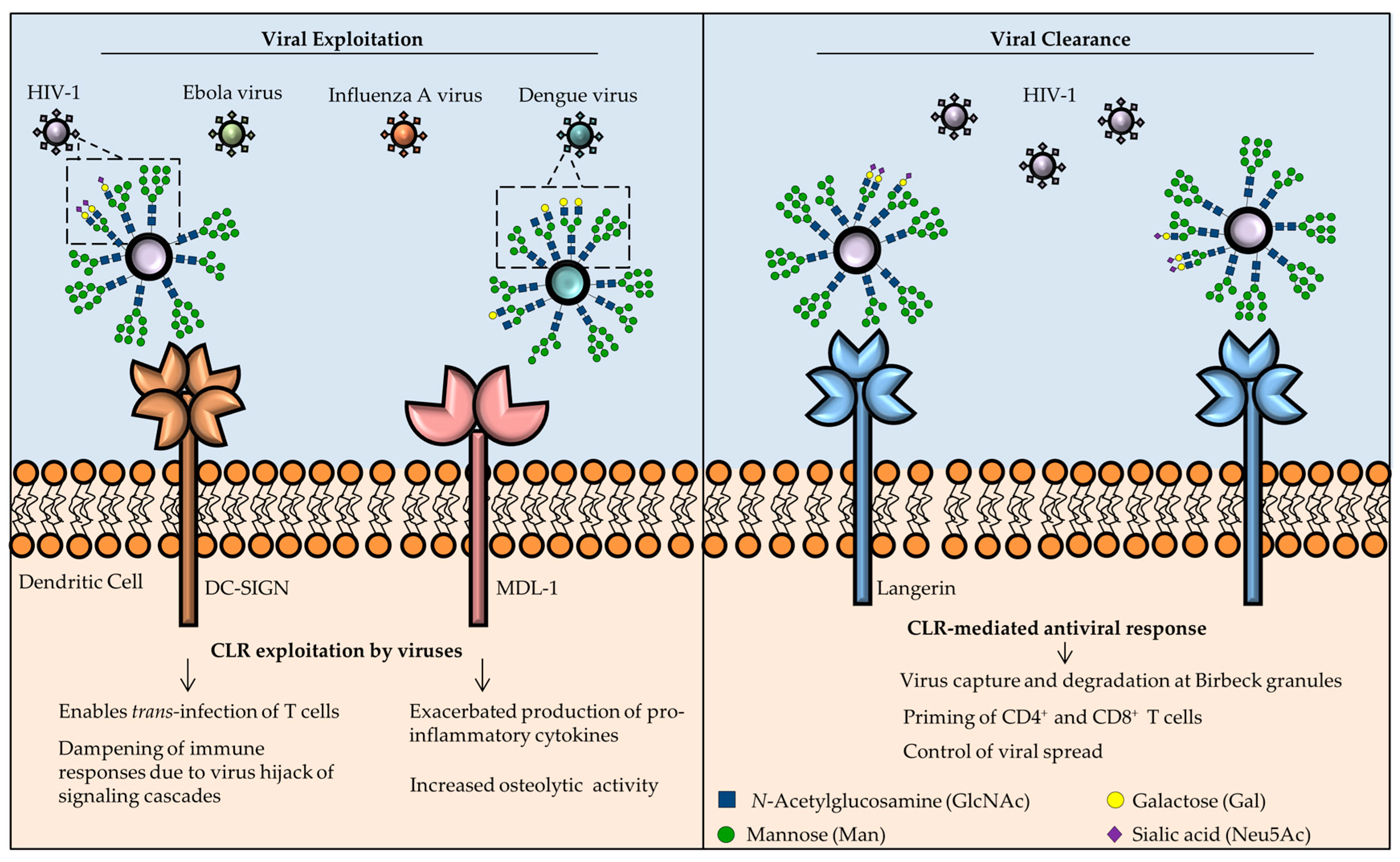
3.1. DC-SIGN
3.2. L-SIGN
3.3. LSECtin
3.4. Langerin
3.5. Dendritic Cell Immunoreceptor
3.6. MDL-1
3.7. Macrophage Mannose Receptor
3.8. Macrophage Galactose-Type Lectin
4. Multivalent Glycoconjugates as Antivirals
5. Conclusion and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Murphy, K.; Travers, P.; Walport, M.; Janeway, C. Janeway’s Immunobiology; Garland Science: New York, NY, USA, 2012. [Google Scholar]
- Beutler, B. Innate immunity: An overview. Mol. Immunol. 2004, 40, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T.D.; Lamkanfi, M.; Nunez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.-M.; Gale, M., Jr. Immune Signaling by RIG-I-like Receptors. Immunity 2016, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; Ohba, Y.; Taniguchi, T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Dambuza, I.M.; Brown, G.D. C-type lectins in immunity: Recent developments. Curr. Opin. Immunol. 2015, 32, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.J.; Sancho, D.; Slack, E.C.; LeibundGut-Landmann, S.; Sousa, C.R. e Myeloid C-type lectins in innate immunity. Nat. Immunol. 2006, 7, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Dam, T.K.; Fred Brewer, C. Lectins as pattern recognition molecules: The effects of epitope density in innate immunity. Glycobiology 2009, 20, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Sancho, D.; Reis e Sousa, C. Signaling by Myeloid C-Type Lectin Receptors in Immunity and Homeostasis. Annu. Rev. Immunol. 2012, 30, 491–529. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, S.J.; Garcia-Vallejo, J.J.; van Kooyk, Y. Dendritic cells and C-type lectin receptors: Coupling innate to adaptive immune responses. Immunol. Cell Biol. 2008, 86, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, D.; Gewurz, B.E.; Furman, M.H.; Schust, D.J.; Ploegh, H.L. Viral Subversion of the Immune System. Annu. Rev. Immunol. 2000, 18, 861–926. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Hart, G.; Stanley, P.; Aebi, M.; Darvill, A.; Kinoshita, T.; Packer, N.H. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- Taylor, M.E.; Drickamer, K. Introduction to Glycobiology; OUP Oxford: Toronto, Canada, 2011. [Google Scholar]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.J.; Milner, J.D.; Rosenzweig, S.D. Glycans Instructing Immunity: The Emerging Role of Altered Glycosylation in Clinical Immunology. Front. Pediatr. 2015, 3, 54. [Google Scholar] [CrossRef] [PubMed]
- Wopereis, S.; Lefeber, D.J.; Morava, É.; Wevers, R.A. Mechanisms in Protein O-Glycan Biosynthesis and Clinical and Molecular Aspects of Protein O-Glycan Biosynthesis Defects: A Review. Clin. Chem. 2006, 52, 574 LP–600 LP. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; Varki, A. Chemical Diversity in the Sialic Acids and Related α-Keto Acids: An Evolutionary Perspective. Chem. Rev. 2002, 102, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Micro 2014, 12, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Van Breedam, W.; Pöhlmann, S.; Favoreel, H.W.; de Groot, R.J.; Nauwynck, H.J. Bitter-sweet symphony: Glycan–lectin interactions in virus biology. FEMS Microbiol. Rev. 2014, 38, 598–632. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, K.; Chandrasekaran, A.; Srinivasan, A.; Raman, R.; Sasisekharan, V.; Sasisekharan, R. Glycans as receptors for influenza pathogenesis. Glycoconj. J. 2010, 27, 561–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Goffard, A.; Callens, N.; Bartosch, B.; Wychowski, C.; Cosset, F.-L.; Montpellier, C.; Dubuisson, J. Role of N-Linked Glycans in the Functions of Hepatitis C Virus Envelope Glycoproteins. J. Virol. 2005, 79, 8400–8409. [Google Scholar] [CrossRef] [PubMed]
- Beyene, A.; Basu, A.; Meyer, K.; Ray, R. Influence of N-linked glycans on intracellular transport of hepatitis C virus E1 chimeric glycoprotein and its role in pseudotype virus infectivity. Virology 2004, 324, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Doores, K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015, 282, 4679–4691. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K.; Taylor, M.E. Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 2015, 34, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef] [PubMed]
- Kiyotake, R.; Oh-hora, M.; Ishikawa, E.; Miyamoto, T.; Ishibashi, T.; Yamasaki, S. Human Mincle Binds to Cholesterol Crystals and Triggers Innate Immune Responses. J. Biol. Chem. 2015, 290, 25322–25332. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Castiñeiras-Vilariño, M.; Höckendorf, U.; Hannesschläger, N.; Lemeer, S.; Kupka, D.; Meyermann, S.; Lech, M.; Anders, H.-J.; Kuster, B.; et al. Clec12a Is an Inhibitory Receptor for Uric Acid Crystals that Regulates Inflammation in Response to Cell Death. Immunity 2016, 40, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Hemmi, H. Dendritic Cells: Translating Innate to Adaptive Immunity. In From Innate Immunity to Immunological Memory; Pulendran, B., Ahmed, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 17–58. [Google Scholar]
- Mayer, S.; Raulf, M.-K.; Lepenies, B. C-type lectins: Their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 2017, 147, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Osorio, F.; Reis e Sousa, C. Myeloid C-type Lectin Receptors in Pathogen Recognition and Host Defense. Immunity 2011, 34, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Kerrigan, A.M.; Brown, G.D. Syk-coupled C-type lectins in immunity. Trends Immunol. 2011, 32, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Ma, S.; Zhou, H.; Zang, A.; Fang, Y.; Li, T.; Shi, H.; Liu, M.; Du, M.; Taylor, P.R.; et al. Tyrosine phosphatase SHP-2 mediates C-type lectin receptor-induced activation of the kinase Syk and anti-fungal TH17 responses. Nat. Immunol. 2015, 16, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.P.; Tarr, A.W. Human lectins and their roles in viral infections. Molecules 2015, 20, 2229–2271. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Pang, X.; Liu, T.; Ning, Z.; Cheng, G. The roles of direct recognition by animal lectins in antiviral immunity and viral pathogenesis. Molecules 2015, 20, 2272–2295. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.F.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a Novel Dendritic Cell–Specific ICAM-3 Receptor that Supports Primary Immune Responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Mitchell, D.A.; Fadden, A.J.; Drickamer, K. A Novel Mechanism of Carbohydrate Recognition by the C-type Lectins DC-SIGN and DC-SIGNR: Subunit Organization and Binding to Multivalent Ligands. J. Biol. Chem. 2001, 276, 28939–28945. [Google Scholar] [CrossRef] [PubMed]
- Khoo, U.-S.; Chan, K.Y.K.; Chan, V.S.F.; Lin, C.L.S. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. 2008, 86, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, D.; Haak, S.; Sisirak, V.; Reizis, B. The role of dendritic cells in autoimmunity. Nat. Rev. Immunol. 2013, 13, 566–577. [Google Scholar] [CrossRef] [PubMed]
- van Liempt, E.; Bank, C.M.C.; Mehta, P.; Garcı´a-Vallejo, J.J.; Kawar, Z.S.; Geyer, R.; Alvarez, R.A.; Cummings, R.D.; Kooyk, Y.; van Die, I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006, 580, 6123–6131. [Google Scholar] [CrossRef] [PubMed]
- Iborra, S.; Sancho, D. Signalling versatility following self and non-self sensing by myeloid C-type lectin receptors. Immunobiology 2015, 220, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.A.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.L.; et al. DC-SIGN (CD209) Mediates Dengue Virus Infection of Human Dendritic Cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Gramberg, T.; Simmons, G.; Möller, P.; Rennekamp, A.J.; Krumbiegel, M.; Geier, M.; Eisemann, J.; Turza, N.; Saunier, B.; et al. DC-SIGN and DC-SIGNR Interact with the Glycoprotein of Marburg Virus and the S Protein of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2004, 78, 12090–12095. [Google Scholar] [CrossRef] [PubMed]
- Baribaud, F.; Pöhlmann, S.; Leslie, G.; Mortari, F.; Doms, R.W. Quantitative Expression and Virus Transmission Analysis of DC-SIGN on Monocyte-Derived Dendritic Cells. J. Virol. 2002, 76, 9135–9142. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Simmons, G.; Pöhlmann, S.; Baribaud, F.; Ni, H.; Leslie, G.J.; Haggarty, B.S.; Bates, P.; Weissman, D.; Hoxie, J.A.; et al. Differential N-Linked Glycosylation of Human Immunodeficiency Virus and Ebola Virus Envelope Glycoproteins Modulates Interactions with DC-SIGN and DC-SIGNR. J. Virol. 2003, 77, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Möller, P.; Hanna, S.L.; Harrer, T.; Eisemann, J.; Steinkasserer, A.; Becker, S.; Baribaud, F.; Pöhlmann, S. Analysis of the Interaction of Ebola Virus Glycoprotein with DC-SIGN (Dendritic Cell—Specific Intercellular Adhesion Molecule 3—Grabbing Nonintegrin) and Its Homologue DC-SIGNR. J. Infect. Dis. 2007, 196, S237–S246. [Google Scholar] [CrossRef] [PubMed]
- van Kooyk, Y.; Geijtenbeek, T.B.H. DC-SIGN: Escape mechanism for pathogens. Nat. Rev. Immunol. 2003, 3, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.F.; Middel, J.; Cornelissen, I.L.M.H.; Nottet, H.S.L.M.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a Dendritic Cell–Specific HIV-1-Binding Protein that Enhances trans-Infection of T Cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef]
- Haase, A.T. Targeting early infection to prevent HIV-1 mucosal transmission. Nature 2010, 464, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Gringhuis, S.I.; van der Vlist, M.; van den Berg, L.M.; den Dunnen, J.; Litjens, M.; Geijtenbeek, T.B.H. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat. Immunol. 2010, 11, 419–426. [Google Scholar] [PubMed]
- Miyauchi, K.; Kim, Y.; Latinovic, O.; Morozov, V.; Melikyan, G.B. HIV Enters Cells via Endocytosis and Dynamin-Dependent Fusion with Endosomes. Cell 2009, 137, 433–444. [Google Scholar] [CrossRef] [PubMed]
- van Montfort, T.; Eggink, D.; Boot, M.; Tuen, M.; Hioe, C.E.; Berkhout, B.; Sanders, R.W. HIV-1 N-Glycan Composition Governs a Balance between Dendritic Cell-Mediated Viral Transmission and Antigen Presentation. J. Immunol. 2011, 187, 4676–4685. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D. Dendritic Cells and HIV-1 Trans-Infection. Viruses 2010, 2, 1704–1717. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Wu, L.; Bohks, S.M.; KewalRamani, V.N.; Unutmaz, D.; Hope, T.J. Recruitment of HIV and Its Receptors to Dendritic Cell-T Cell Junctions. Science 2003, 300, 1295–1297. [Google Scholar] [CrossRef] [PubMed]
- Hodges, A. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat. Immunol. 2007, 8, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hwang, S.-L.; Chan, V.S.F.; Chung, N.P.Y.; Wang, S.-R.; Li, Z.; Ma, J.; Lin, C.-W.; Hsieh, Y.-J.; Chang, K.-P.; et al. Binding of HIV-1 gp120 to DC-SIGN Promotes ASK-1-Dependent Activation-Induced Apoptosis of Human Dendritic Cells. PLoS Pathog. 2013, 9, e1003100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozach, P.-Y.; Burleigh, L.; Staropoli, I.; Amara, A. The C Type Lectins DC-SIGN and L-SIGN. In Glycovirology Protocols SE - 4; Sugrue, R., Ed.; Methods in Molecular Biology; Humana Press: New Jersey, USA, 2007; Volume 379, pp. 51–68. [Google Scholar]
- Celerino da Silva, R.; Segat, L.; Crovella, S. Role of DC-SIGN and L-SIGN receptors in HIV-1 vertical transmission. Hum. Immunol. 2011, 72, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, I.S.; Lekkerkerker, A.N.; Depla, E.; Bosman, F.; Musters, R.J.P.; Depraetere, S.; van Kooyk, Y.; Geijtenbeek, T.B.H. Hepatitis C Virus Targets DC-SIGN and L-SIGN To Escape Lysosomal Degradation. J. Virol. 2004, 78, 8322–8332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, G.; Reeves, J.D.; Grogan, C.C.; Vandenberghe, L.H.; Baribaud, F.; Whitbeck, J.C.; Burke, E.; Buchmeier, M.J.; Soilleux, E.J.; Riley, J.L.; et al. DC-SIGN and DC-SIGNR Bind Ebola Glycoproteins and Enhance Infection of Macrophages and Endothelial Cells. Virology 2003, 305, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Nangle, E.M.; Smith, M.S.; Yurochko, A.D.; Ryman, K.D. DC-SIGN and L-SIGN Can Act as Attachment Receptors for Alphaviruses and Distinguish between Mosquito Cell- and Mammalian Cell-Derived Viruses. J. Virol. 2003, 77, 12022–12032. [Google Scholar] [CrossRef] [PubMed]
- Op den Brouw, M.L.; De Jong, M.A.W.P.; Ludwig, I.S.; Van Der Molen, R.G.; Janssen, H.L.A.; Geijtenbeek, T.B.H.; Woltman, A.M. Branched oligosaccharide structures on HBV prevent interaction with both DC-SIGN and L-SIGN. J. Viral Hepat. 2008, 15, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Han, D.P.; Lohani, M.; Cho, M.W. Specific Asparagine-Linked Glycosylation Sites Are Critical for DC-SIGN- and L-SIGN-Mediated Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2007, 81, 12029–12039. [Google Scholar] [CrossRef] [PubMed]
- Brauburger, K.; Hume, A.J.; Mühlberger, E.; Olejnik, J. Forty-Five Years of Marburg Virus Research. Viruses 2012, 4, 1878–1927. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.W.; Nguyen, H.-Y.; Hanna, S.L.; Sánchez, M.D.; Doms, R.W.; Pierson, T.C. West Nile Virus Discriminates between DC-SIGN and DC-SIGNR for Cellular Attachment and Infection. J. Virol. 2006, 80, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Léger, P.; Tetard, M.; Youness, B.; Cordes, N.; Rouxel, R.N.; Flamand, M.; Lozach, P.-Y. Differential Use of the C-Type Lectins L-SIGN and DC-SIGN for Phlebovirus Endocytosis. Traffic 2016, 17, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.; Roosendahl, P.; Ng, W.C.; Brooks, A.G.; Reading, P.C.; Londrigan, S.L. Endocytic function is critical for influenza A virus infection via DC-SIGN and L-SIGN. Sci. Rep. 2016, 6, 19428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ren, S.; Zuo, Y. DC-SIGN, DC-SIGNR and LSECtin: C-Type Lectins for Infection. Int. Rev. Immunol. 2014, 33, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, L.; Zhang, G.; Wei, H.; Cui, Y.; Guo, L.; Gou, Z.; Chen, X.; Jiang, D.; Zhu, Y.; et al. Characterization of a Novel C-type Lectin-like Gene, LSECtin: Demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J. Biol. Chem. 2004, 279, 18748–18758. [Google Scholar] [CrossRef] [PubMed]
- Gramberg, T.; Hofmann, H.; Möller, P.; Lalor, P.F.; Marzi, A.; Geier, M.; Krumbiegel, M.; Winkler, T.; Kirchhoff, F.; Adams, D.H.; et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 2005, 340, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Gramberg, T.; Soilleux, E.; Fisch, T.; Lalor, P.F.; Hofmann, H.; Wheeldon, S.; Cotterill, A.; Wegele, A.; Winkler, T.; Adams, D.H.; Pöhlmann, S. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology 2008, 373, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Soto, A.; Aragoneses-Fenoll, L.; Martin-Gayo, E.; Martinez-Prats, L.; Colmenares, M.; Naranjo-Gomez, M.; Borras, F.E.; Munoz, P.; Zubiaur, M.; Toribio, M.L.; et al. The DC-SIGN–related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood 2007, 109, 5337–5345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L.; Yang, J.; Liu, W.; Tang, X.; Chen, J.; Zhao, D.; Wang, M.; Xu, F.; Lu, Y.; Liu, B.; et al. Liver Sinusoidal Endothelial Cell Lectin, LSECtin, Negatively Regulates Hepatic T-Cell Immune Response. Gastroenterology 2009, 137, 1498–1508.e5. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Kwota, Z.; Sun, J. Intrahepatic regulation of antiviral T cell responses at initial stages of viral infection. Int. Immunopharmacol. 2016, 39, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, M.; Wang, X.; Zhao, D.; Liu, D.; Liu, J.; Chen, P.-J.; Yang, D.; He, F.; Tang, L. Liver Sinusoidal Endothelial Cell Lectin Inhibits CTL-Dependent Virus Clearance in Mouse Models of Viral Hepatitis. J. Immunol. 2013, 190, 4185–4195. [Google Scholar] [CrossRef] [PubMed]
- Stoitzner, P.; Romani, N. Langerin, the “Catcher in the Rye”: An important receptor for pathogens on Langerhans cells. Eur. J. Immunol. 2011, 41, 2526–2529. [Google Scholar] [CrossRef] [PubMed]
- Valladeau, J.; Ravel, O.; Dezutter-Dambuyant, C.; Moore, K.; Kleijmeer, M.; Liu, Y.; Duvert-Frances, V.; Vincent, C.; Schmitt, D.; Davoust, J.; et al. Langerin, a Novel C-Type Lectin Specific to Langerhans Cells, Is an Endocytic Receptor that Induces the Formation of Birbeck Granules. Immunity 2000, 12, 71–81. [Google Scholar] [CrossRef]
- Stambach, N.S.; Taylor, M.E. Characterization of carbohydrate recognition by langerin, a C-type lectin of Langerhans cells. Glycobiol. 2003, 13, 401–410. [Google Scholar] [CrossRef] [PubMed]
- van der Vlist, M.; Geijtenbeek, T.B.H. Langerin functions as an antiviral receptor on Langerhans cells. Immunol. Cell Biol. 2010, 88, 410–415. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.A.W.P.; Vriend, L.E.M.; Theelen, B.; Taylor, M.E.; Fluitsma, D.; Boekhout, T.; Geijtenbeek, T.B.H. C-type lectin Langerin is a β-glucan receptor on human Langerhans cells that recognizes opportunistic and pathogenic fungi. Mol. Immunol. 2010, 47, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.L.; Abendroth, A.; Jones, C.; Nasr, N.; Turville, S. Viruses and Langerhans cells. Immunol. Cell Biol. 2010, 88, 416–423. [Google Scholar] [CrossRef] [PubMed]
- de Witte, L.; Nabatov, A.; Pion, M.; Fluitsma, D.; de Jong, M.A.W.P.; de Gruijl, T.; Piguet, V.; van Kooyk, Y.; Geijtenbeek, T.B.H. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 2007, 13, 367–371. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.A.W.P.; de Witte, L.; Oudhoff, M.J.; Gringhuis, S.I.; Gallay, P.; Geijtenbeek, T.B.H. TNF-α and TLR agonists increase susceptibility to HIV-1 transmission by human Langerhans cells ex vivo. J. Clin. Invest. 2008, 118, 3440–3452. [Google Scholar] [CrossRef] [PubMed]
- van der Vlist, M.; de Witte, L.; de Vries, R.D.; Litjens, M.; de Jong, M.A.W.P.; Fluitsma, D.; de Swart, R.L.; Geijtenbeek, T.B.H. Human Langerhans cells capture measles virus through Langerin and present viral antigens to CD4+ T cells but are incapable of cross-presentation. Eur. J. Immunol. 2011, 41, 2619–2631. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.C.; Londrigan, S.L.; Nasr, N.; Cunningham, A.L.; Turville, S.; Brooks, A.G.; Reading, P.C. The C-type lectin langerin functions as a receptor for attachment and infectious entry of influenza A virus. J. Virol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Bloem, K.; Vuist, I.M.; van den Berk, M.; Klaver, E.J.; van Die, I.; Knippels, L.M.J.; Garssen, J.; García-Vallejo, J.J.; van Vliet, S.J.; van Kooyk, Y. DCIR interacts with ligands from both endogenous and pathogenic origin. Immunol. Lett. 2014, 158, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.A.; Imbeault, M.; Gilbert, C.; Tremblay, M.J. HIV-1 Induces DCIR Expression in CD4+ T Cells. PLoS Pathog. 2010, 6, e1001188. [Google Scholar] [CrossRef] [PubMed]
- Bloem, K.; Vuist, I.M.; van der Plas, A.-J.; Knippels, L.M.J.; Garssen, J.; García-Vallejo, J.J.; van Vliet, S.J.; van Kooyk, Y. Ligand Binding and Signaling of Dendritic Cell Immunoreceptor (DCIR) Is Modulated by the Glycosylation of the Carbohydrate Recognition Domain. PLoS ONE 2013, 8, e66266. [Google Scholar] [CrossRef] [PubMed]
- Fujikado, N.; Saijo, S.; Yonezawa, T.; Shimamori, K.; Ishii, A.; Sugai, S.; Kotaki, H.; Sudo, K.; Nose, M.; Iwakura, Y. Dcir deficiency causes development of autoimmune diseases in mice due to excess expansion of dendritic cells. Nat. Med. 2008, 14, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Wentrup, F.; Benitez-Ribas, D.; Tacken, P.J.; Punt, C.J.A.; Figdor, C.G.; de Vries, I.J.M.; Adema, G.J. Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-α production. Blood 2008, 111, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Klechevsky, E.; Flamar, A.-L.; Cao, Y.; Blanck, J.-P.; Liu, M.; O’Bar, A.; Agouna-Deciat, O.; Klucar, P.; Thompson-Snipes, L.; Zurawski, S.; et al. Cross-priming CD8+ T cells by targeting antigens to human dendritic cells through DCIR. Blood 2010, 116, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.A.; Gilbert, C.; Richard, M.; Beaulieu, A.D.; Tremblay, M.J. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood 2008, 112, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.A.; Barabé, F.; Gilbert, C.; Tremblay, M.J. DCIR-mediated enhancement of HIV-1 infection requires the ITIM-associated signal transduction pathway. Blood 2011, 117, 6589–6599. [Google Scholar] [CrossRef] [PubMed]
- Flamar, A.-L.; Contreras, V.; Zurawski, S.; Montes, M.; Dereuddre-Bosquet, N.; Martinon, F.; Banchereau, J.; Le Grand, R.; Zurawski, G.; Levy, Y. Delivering HIV Gagp24 to DCIR Induces Strong Antibody Responses In Vivo. PLoS ONE 2015, 10, e0135513. [Google Scholar] [CrossRef] [PubMed]
- Long, K.M.; Whitmore, A.C.; Ferris, M.T.; Sempowski, G.D.; McGee, C.; Trollinger, B.; Gunn, B.; Heise, M.T. Dendritic Cell Immunoreceptor Regulates Chikungunya Virus Pathogenesis in Mice. J. Virol. 2013, 87, 5697–5706. [Google Scholar] [CrossRef] [PubMed]
- Maglinao, M.; Klopfleisch, R.; Seeberger, P.H.; Lepenies, B. The C-Type Lectin Receptor DCIR Is Crucial for the Development of Experimental Cerebral Malaria. J. Immunol. 2013, 191, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
- Bakker, A.B.H.; Baker, E.; Sutherland, G.R.; Phillips, J.H.; Lanier, L.L. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc. Natl. Acad. Sci. USA 1999, 96, 9792–9796. [Google Scholar] [CrossRef] [PubMed]
- Yim, D.; Jie, H.-B.; Sotiriadis, J.; Kim, Y.-S.; Kim, Y.B. Molecular Cloning and Expression Pattern of Porcine Myeloid DAP12-Associating Lectin-1. Cell. Immunol. 2001, 209, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L.; Bakker, A.B.H. The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol. Today 2000, 21, 611–614. [Google Scholar] [CrossRef]
- Watson, A.A.; Lebedev, A.A.; Hall, B.A.; Fenton-May, A.E.; Vagin, A.A.; Dejnirattisai, W.; Felce, J.; Mongkolsapaya, J.; Palma, A.S.; Liu, Y.; et al. Structural Flexibility of the Macrophage Dengue Virus Receptor CLEC5A: Implications for Ligand Binding and Signaling. J. Biol. Chem. 2011, 286, 24208–24218. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.A.; O’Callaghan, C.A. Crystallization and X-ray diffraction analysis of human CLEC5A (MDL-1), a dengue virus receptor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-T.; Lin, Y.-L.; Huang, M.-T.; Wu, M.-F.; Cheng, S.-C.; Lei, H.-Y.; Lee, C.-K.; Chiou, T.-W.; Wong, C.-H.; Hsieh, S.-L. CLEC5A is critical for dengue-virus-induced lethal disease. Nature 2008, 453, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-T.; Liu, R.-S.; Wu, M.-F.; Lin, Y.-L.; Chen, S.-Y.; Tan, D.T.-W.; Chou, T.-Y.; Tsai, I.-S.; Li, L.; Hsieh, S.-L. CLEC5A Regulates Japanese Encephalitis Virus-Induced Neuroinflammation and Lethality. PLoS Pathog. 2012, 8, e1002655. [Google Scholar] [CrossRef] [PubMed]
- Teng, O.; Chen, S.-T.; Hsu, T.-L.; Sia, S.F.; Cole, S.; Valkenburg, S.A.; Hsu, T.-Y.; Zheng, J.T.; Tu, W.; Bruzzone, R.; Peiris, J.S.M.; Hsieh, S.-L.; Yen, H.-L. CLEC5A-mediated enhancement of the inflammatory response in myeloid cells contributes to influenza pathogenicity in vivo. J. Virol. 2016. [Google Scholar]
- Khetarpal, N.; Khanna, I. Dengue Fever: Causes, Complications, and Vaccine Strategies. J. Immunol. Res. 2016, 2016, 6803098. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-F.; Chen, S.-T.; Yang, A.-H.; Lin, W.-W.; Lin, Y.-L.; Chen, N.-J.; Tsai, I.-S.; Li, L.; Hsieh, S.-L. CLEC5A is critical for dengue virus–induced inflammasome activation in human macrophages. Blood 2013, 121, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Cardosa, M.J.; Guzman, M.G. Of cascades and perfect storms: The immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF//DSS). Immunol. Cell Biol. 2006, 85, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-F.; Chen, S.-T.; Hsieh, S.-L. Distinct regulation of dengue virus-induced inflammasome activation in human macrophage subsets. J. Biomed. Sci. 2013, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-L.; Lin, Y.-S.; Chen, C.-L.; Tsai, T.-T.; Tsai, C.-C.; Wu, Y.-W.; Ou, Y.-D.; Chu, Y.-Y.; Wang, J.-M.; Yu, C.-Y.; et al. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF-α production by mononuclear phagocytes. Sci. Rep. 2016, 6, 32000. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Kikuchi, Y.; Aoki, N.; Endo, S.; Maeda, T.; Sugahara-Tobinai, A.; Fujimura, S.; Nakamura, A.; Kumanogoh, A.; Colonna, M.; Takai, T. Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc. Natl. Acad. Sci. USA 2009, 106, 4816–4821. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-L.; Chen, S.-T.; Liu, R.-S.; Chen, Y.-H.; Lin, C.-Y.; Huang, C.-H.; Shu, P.-Y.; Liao, C.-L.; Hsieh, S.-L. CLEC5A is critical for dengue virus-induced osteoclast activation and bone homeostasis. J. Mol. Med. 2016, 94, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pomares, L. The mannose receptor. J. Leukoc. Biol. 2012, 92, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Bernhard, O.K.; Turville, S.G.; Harman, A.N.; Wilkinson, J.; Cunningham, A.L. Oligomerization of the Macrophage Mannose Receptor Enhances gp120-mediated Binding of HIV-1. J. Biol. Chem. 2009, 284, 11027–11038. [Google Scholar] [CrossRef] [PubMed]
- Gazi, U.; Martinez-Pomares, L. Influence of the mannose receptor in host immune responses. Immunobiology 2009, 214, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.G.; Hildreth, J.E.K. Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur. J. Immunol. 2003, 33, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; deWet, B.J.M.; Martinez-Pomares, L.; Radcliffe, C.M.; Dwek, R.A.; Rudd, P.M.; Gordon, S. The Mannose Receptor Mediates Dengue Virus Infection of Macrophages. PLoS Pathog. 2008, 4, e17. [Google Scholar] [CrossRef]
- Upham, J.P.; Pickett, D.; Irimura, T.; Anders, E.M.; Reading, P.C. Macrophage Receptors for Influenza A Virus: Role of the Macrophage Galactose-Type Lectin and Mannose Receptor in Viral Entry. J. Virol. 2010, 84, 3730–3737. [Google Scholar] [CrossRef] [PubMed]
- Reading, P.C.; Miller, J.L.; Anders, E.M. Involvement of the Mannose Receptor in Infection of Macrophages by Influenza Virus. J. Virol. 2000, 74, 5190–5197. [Google Scholar] [CrossRef] [PubMed]
- Op den Brouw, M.L.; Binda, R.S.; Geijtenbeek, T.B.H.; Janssen, H.L.A.; Woltman, A.M. The mannose receptor acts as hepatitis B virus surface antigen receptor mediating interaction with intrahepatic dendritic cells. Virology 2009, 393, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Zizzari, I.G.; Napoletano, C.; Battisti, F.; Rahimi, H.; Caponnetto, S.; Pierelli, L.; Nuti, M.; Rughetti, A. MGL Receptor and Immunity: When the Ligand Can Make the Difference. J. Immunol. Res. 2015, 2015, 450695. [Google Scholar] [CrossRef] [PubMed]
- Usami, K.; Matsuno, K.; Igarashi, M.; Denda-Nagai, K.; Takada, A.; Irimura, T. Involvement of viral envelope GP2 in Ebola virus entry into cells expressing the macrophage galactose-type C-type lectin. Biochem. Biophys. Res. Commun. 2011, 407, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Fujioka, K.; Tsuiji, M.; Morikawa, A.; Higashi, N.; Ebihara, H.; Kobasa, D.; Feldmann, H.; Irimura, T.; Kawaoka, Y. Human Macrophage C-Type Lectin Specific for Galactose and N-Acetylgalactosamine Promotes Filovirus Entry. J. Virol. 2004, 78, 2943–2947. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K.; Kishida, N.; Usami, K.; Igarashi, M.; Yoshida, R.; Nakayama, E.; Shimojima, M.; Feldmann, H.; Irimura, T.; Kawaoka, Y.; et al. Different Potential of C-Type Lectin-Mediated Entry between Marburg Virus Strains. J. Virol. 2010, 84, 5140–5147. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.C.; Liong, S.; Tate, M.D.; Irimura, T.; Denda-Nagai, K.; Brooks, A.G.; Londrigan, S.L.; Reading, P.C. The Macrophage Galactose-Type Lectin Can Function as an Attachment and Entry Receptor for Influenza Virus. J. Virol. 2014, 88, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, A.; Jiménez-Barbero, J.; Casnati, A.; De Castro, C.; Darbre, T.; Fieschi, F.; Finne, J.; Funken, H.; Jaeger, K.-E.; Lahmann, M.; et al. Multivalent glycoconjugates as anti-pathogenic agents. Chem. Soc. Rev. 2013, 42, 4709–4727. [Google Scholar] [CrossRef] [PubMed]
- Lepenies, B.; Lee, J.; Sonkaria, S. Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv. Drug Deliv. Rev. 2013, 65, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Johannssen, T.; Lepenies, B. Glycan-Based Cell Targeting To Modulate Immune Responses. Trends Biotechnol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, L.L.; Splain, R.A. Chemical Approaches to Glycobiology. Annu. Rev. Biochem. 2010, 79, 619–653. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.D.; Unger, W.J.; Storm, G.; van Kooyk, Y.; Mastrobattista, E. Targeting tumor antigens to dendritic cells using particulate carriers. J. Control. Release 2012, 161, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Becer, C.R. The glycopolymer code: Synthesis of glycopolymers and multivalent carbohydrate-lectin interactions. Macromol. Rapid Commun. 2012, 33, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Berzi, A.; Reina, J.J.; Ottria, R.; Sutkeviciute, I.; Antonazzo, P.; Sanchez-Navarro, M.; Chabrol, E.; Biasin, M.; Trabattoni, D.; Cetin, I.; et al. A glycomimetic compound inhibits DC-SIGN-mediated HIV infection in cellular and cervical explant models. AIDS 2012, 26. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Dane, E.L.; O’Toole, G.A.; Grinstaff, M.W. Exploiting Dendrimer Multivalency to Combat Emerging and Re-Emerging Infectious Diseases. Mol. Pharm. 2012, 9, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.P.; Buffet, K.; Nierengarten, I.; Imberty, A.; Nierengarten, J.-F. Biologically Active Heteroglycoclusters Constructed on a Pillar[5]arene-Containing [2]Rotaxane Scaffold. Chem. – A Eur. J. 2016, 22, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, N. Multivalent ligand presentation as a central concept to study intricate carbohydrate-protein interactions. Chem. Soc. Rev. 2009, 38, 3463–3483. [Google Scholar] [CrossRef] [PubMed]
- Maglinao, M.; Eriksson, M.; Schlegel, M.K.; Zimmermann, S.; Johannssen, T.; Götze, S.; Seeberger, P.H.; Lepenies, B. A platform to screen for C-type lectin receptor-binding carbohydrates and their potential for cell-specific targeting and immune modulation. J. Control. Release 2014, 175, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Brzezicka, K.; Vogel, U.; Serna, S.; Johannssen, T.; Lepenies, B.; Reichardt, N.-C. Influence of Core β-1,2-Xylosylation on Glycoprotein Recognition by Murine C-type Lectin Receptors and Its Impact on Dendritic Cell Targeting. ACS Chem. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Serna, S.; Maglinao, M.; Schlegel, M.K.; Seeberger, P.H.; Reichardt, N.-C.; Lepenies, B. Biological Evaluation of Multivalent Lewis X–MGL-1 Interactions. ChemBioChem 2014, 15, 844–851. [Google Scholar] [CrossRef] [PubMed]
- García-Vallejo, J.J.; Unger, W.W.J.; Kalay, H.; van Kooyk, Y. Glycan-based DC-SIGN targeting to enhance antigen cross-presentation in anticancer vaccines. Oncoimmunology 2013, 2, e23040. [Google Scholar] [CrossRef] [PubMed]
- van Kooyk, Y.; Unger, W.W.J.; Fehres, C.M.; Kalay, H.; García-Vallejo, J.J. Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol. Immunol. 2013, 55, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ávila, O.; Hijazi, K.; Marradi, M.; Clavel, C.; Campion, C.; Kelly, C.; Penadés, S. Gold Manno-Glyconanoparticles: Multivalent Systems to Block HIV-1 gp120 Binding to the Lectin DC-SIGN. Chem. – A Eur. J. 2009, 15, 9874–9888. [Google Scholar] [CrossRef] [PubMed]
- Arnáiz, B.; Martínez-Ávila, O.; Falcon-Perez, J.M.; Penadés, S. Cellular Uptake of Gold Nanoparticles Bearing HIV gp120 Oligomannosides. Bioconjug. Chem. 2012, 23, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ávila, O.; Bedoya, L.M.; Marradi, M.; Clavel, C.; Alcamí, J.; Penadés, S. Multivalent Manno-Glyconanoparticles Inhibit DC-SIGN-Mediated HIV-1 Trans-Infection of Human T Cells. ChemBioChem 2009, 10, 1806–1809. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vallejo, J.J.; Koning, N.; Ambrosini, M.; Kalay, H.; Vuist, I.; Sarrami-Forooshani, R.; Geijtenbeek, T.B.H.; van Kooyk, Y. Glycodendrimers prevent HIV transmission via DC-SIGN on dendritic cells. Int. Immunol. 2013, 25, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Shiao, T.C.; Roy, R. Glycodendrimers as functional antigens and antitumor vaccines. New J. Chem. 2012, 36, 324–339. [Google Scholar] [CrossRef]
- Varga, N.; Sutkeviciute, I.; Ribeiro-Viana, R.; Berzi, A.; Ramdasi, R.; Daghetti, A.; Vettoretti, G.; Amara, A.; Clerici, M.; Rojo, J.; Fieschi, F.; Bernardi, A. A multivalent inhibitor of the DC-SIGN dependent uptake of HIV-1 and Dengue virus. Biomaterials 2014, 35, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Ordanini, S.; Varga, N.; Porkolab, V.; Thepaut, M.; Belvisi, L.; Bertaglia, A.; Palmioli, A.; Berzi, A.; Trabattoni, D.; Clerici, M.; Fieschi, F.; Bernardi, A. Designing nanomolar antagonists of DC-SIGN-mediated HIV infection: Ligand presentation using molecular rods. Chem. Commun. 2015, 51, 3816–3819. [Google Scholar] [CrossRef] [PubMed]
- Sattin, S.; Daghetti, A.; Thépaut, M.; Berzi, A.; Sánchez-Navarro, M.; Tabarani, G.; Rojo, J.; Fieschi, F.; Clerici, M.; Bernardi, A. Inhibition of DC-SIGN-Mediated HIV Infection by a Linear Trimannoside Mimic in a Tetravalent Presentation. ACS Chem. Biol. 2010, 5, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Luczkowiak, J.; Sattin, S.; Sutkevičiu̅tė, I.; Reina, J.J.; Sánchez-Navarro, M.; Thépaut, M.; Martínez-Prats, L.; Daghetti, A.; Fieschi, F.; Delgado, R.; et al. Pseudosaccharide Functionalized Dendrimers as Potent Inhibitors of DC-SIGN Dependent Ebola Pseudotyped Viral Infection. Bioconjug. Chem. 2011, 22, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Nierengarten, J.-F.; Iehl, J.; Oerthel, V.; Holler, M.; Illescas, B.M.; Munoz, A.; Martin, N.; Rojo, J.; Sanchez-Navarro, M.; Cecioni, S.; et al. Fullerene sugar balls. Chem. Commun. 2010, 46, 3860–3862. [Google Scholar] [CrossRef] [PubMed]
- Luczkowiak, J.; Muñoz, A.; Sánchez-Navarro, M.; Ribeiro-Viana, R.; Ginieis, A.; Illescas, B.M.; Martín, N.; Delgado, R.; Rojo, J. Glycofullerenes Inhibit Viral Infection. Biomacromolecules 2013, 14, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, A.; Sigwalt, D.; Illescas, B.M.; Luczkowiak, J.; Rodríguez-Pérez, L.; Nierengarten, I.; Holler, M.; Remy, J.-S.; Buffet, K.; Vincent, S.P.; et al. Synthesis of giant globular multivalent glycofullerenes as potent inhibitors in a model of Ebola virus infection. Nat. Chem. 2016, 8, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Vidal, S. Glycofullerenes: Sweet fullerenes vanquish viruses. Nat. Chem. 2016, 8, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Viana, R.; Sánchez-Navarro, M.; Luczkowiak, J.; Koeppe, J.R.; Delgado, R.; Rojo, J.; Davis, B.G. Virus-like glycodendrinanoparticles displaying quasi-equivalent nested polyvalency upon glycoprotein platforms potently block viral infection. Nat. Commun. 2012, 3, 1303. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.F.; Cummings, R.D. Investigating Virus-Glycan Interactions Using Glycan Microarrays. Curr. Opin. Virol. 2014, 7, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Khatri, K.; Klein, J.A.; White, M.R.; Grant, O.C.; Leymarie, N.; Woods, R.J.; Hartshorn, K.L.; Zaia, J. Integrated Omics and Computational Glycobiology Reveal Structural Basis for Influenza A Virus Glycan Microheterogeneity and Host Interactions. Mol. Cell. Proteom. 2016, 15, 1895–1912. [Google Scholar] [CrossRef] [PubMed]

| CLR | Location | Ligand Specificity | Function | Virus | Target Protein | Reference |
|---|---|---|---|---|---|---|
| DC-SIGN and L-SIGN | Transmembrane | High mannose High fucose | Enable viral infection | HIV | gp120 | [51,54,64] |
| EBOV | GP1 subunit | [51,52] | ||||
| HBV | HBsAg | [68] | ||||
| HCV | Envelope glycoproteins | [65] | ||||
| WNV | E protein or prM protein | [71] | ||||
| DV | E protein | [48] | ||||
| MARV | Envelope glycoproteins | [49] | ||||
| SARS-CoV | S protein | [49] | ||||
| Langerin | Transmembrane | GlcNAc Mannose Fucose | Inhibit viral infection | HIV | gp120 | [85,88] |
| Enable viral infection | MV | F and H protein | [90] | |||
| IAV | HA | [91] | ||||
| MMR | Transmembrane | GlcNAc Mannose Fucose | IAV | HA or NA | [123,124] | |
| HIV | gp120 | [119,121] | ||||
| DV | E protein | [122] | ||||
| HBV | HBsAg | [125] | ||||
| DCIR | Transmembrane | Mannose Fucose | HIV | gp120 | [93,99] | |
| Inhibit viral infection | CHKV | Envelope glycoproteins | [101] | |||
| MDL-1 | Transmembrane | Non-defined | Enable viral infection | JEV | Envelope glycoproteins | [109] |
| DV | Envelope glycoproteins | [108,112,117] | ||||
| IAV | HA | [110] | ||||
| LSECtin | Transmembrane | GlcNAc Mannose Fucose | Enable viral infection | EBOV | GP | [78] |
| HBV | HBsAg | [81] | ||||
| SARS-CoV | Envelope glycoproteins | [76] | ||||
| MGL | Transmembrane | Galactose GalNAc | Enable viral infection | EBOV | GP | [127,128] |
| IAV | HA or NA | [123,130] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, J.T.; Lepenies, B. Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity. Viruses 2017, 9, 59. https://doi.org/10.3390/v9030059
Monteiro JT, Lepenies B. Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity. Viruses. 2017; 9(3):59. https://doi.org/10.3390/v9030059
Chicago/Turabian StyleMonteiro, João T., and Bernd Lepenies. 2017. "Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity" Viruses 9, no. 3: 59. https://doi.org/10.3390/v9030059






