Microbial Natural Product Alternariol 5-O-Methyl Ether Inhibits HIV-1 Integration by Blocking Nuclear Import of the Pre-Integration Complex
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Plasmids and Reagents
2.3. Screening for Anti-HIV Compounds from a Library of Microbial Natural Compounds
2.4. Assay for Measuring the Inhibitory Activity of Compounds on Different HIV-1 Strains
2.5. Cytotoxicity Assay
2.6. Time of Addition Experiment
2.7. Semi-Quantitative Real-Time PCR
2.8. Immunostaining
2.9. Nuclear and Cytoplasmic Fractionation
2.10. Viral Preparation and Infection Assay
2.11. In Vitro Integrase Assay
3. Results
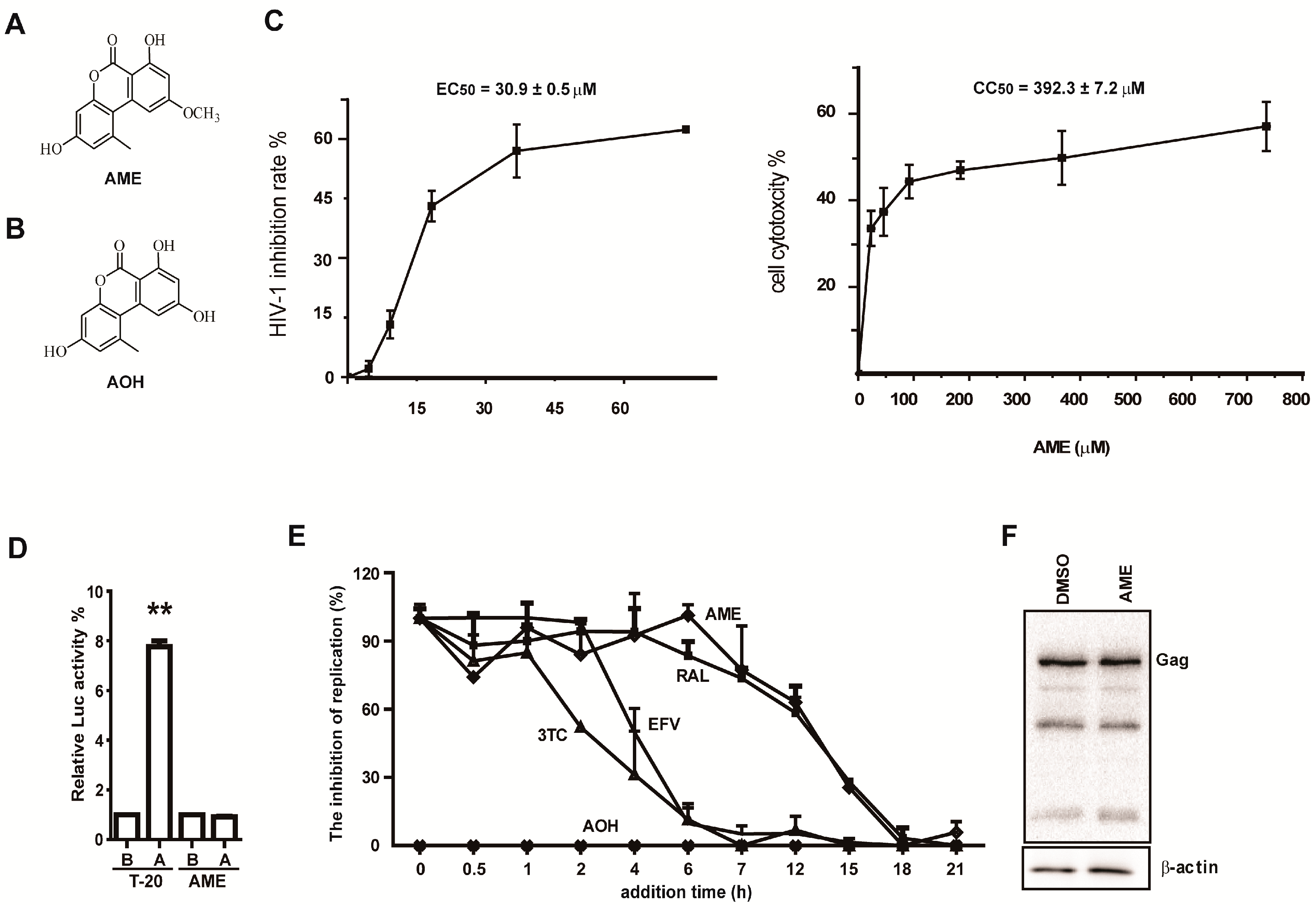
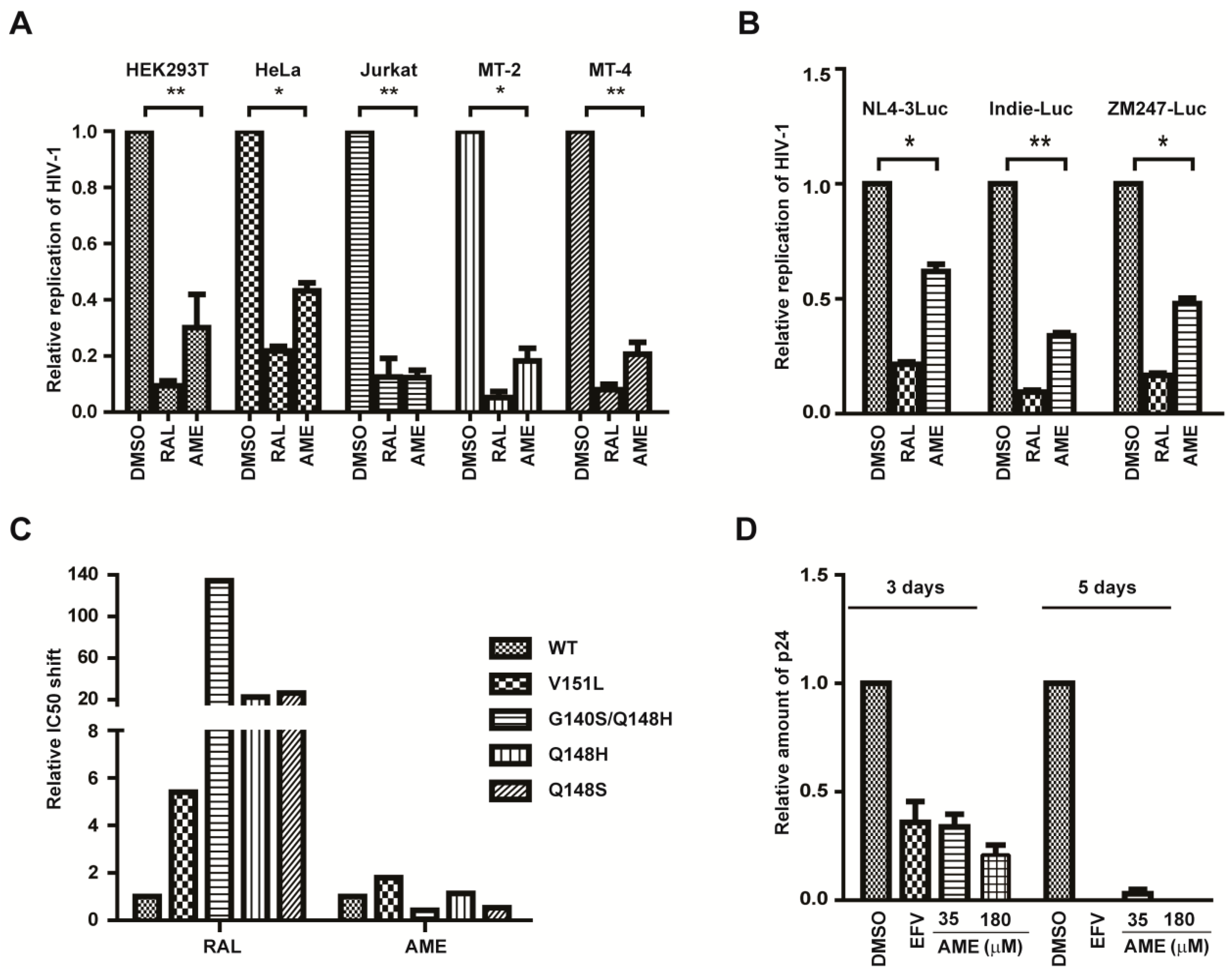
3.1. AME Inhibits HIV Infection
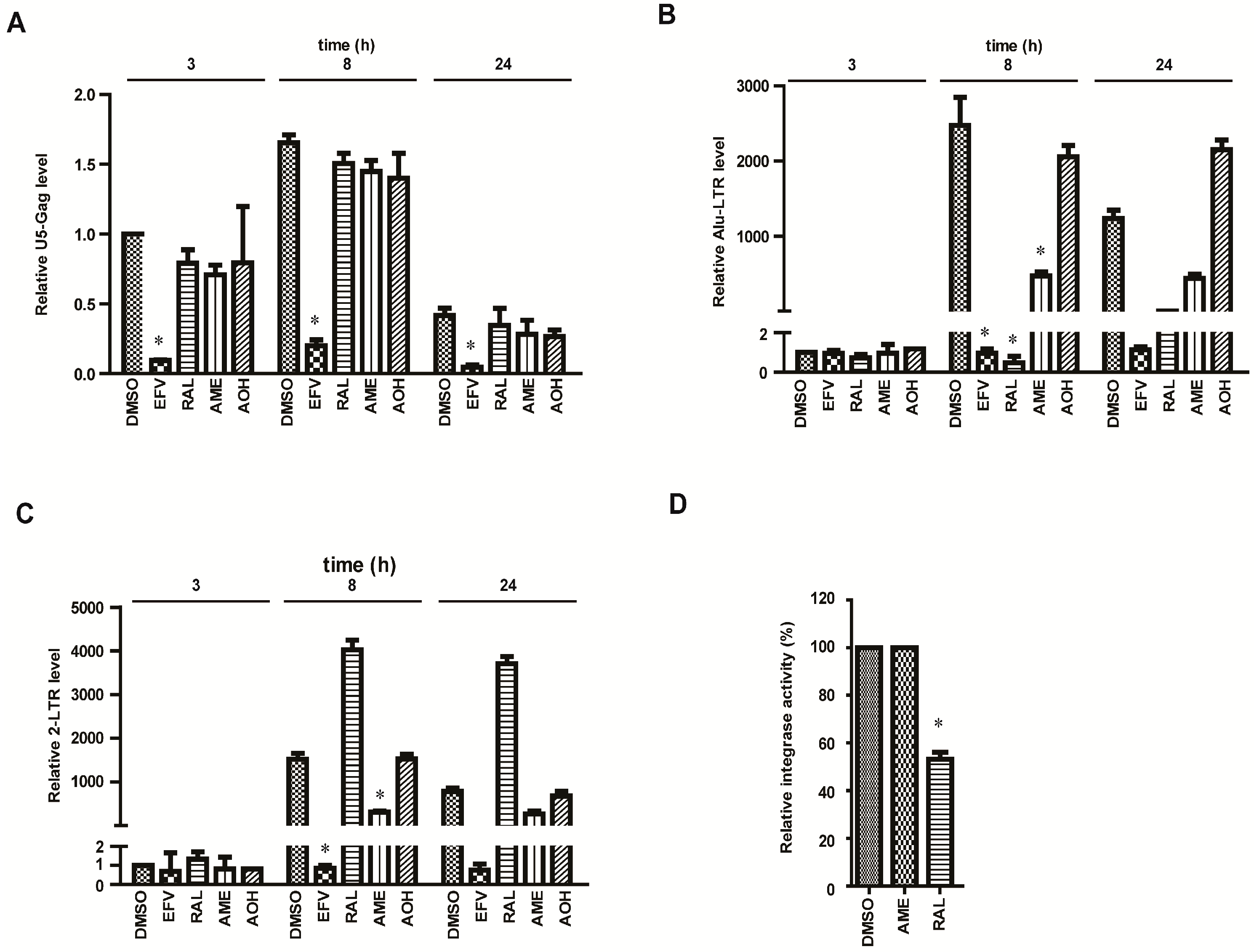
3.2. AME Causes Defective HIV-1 Integration
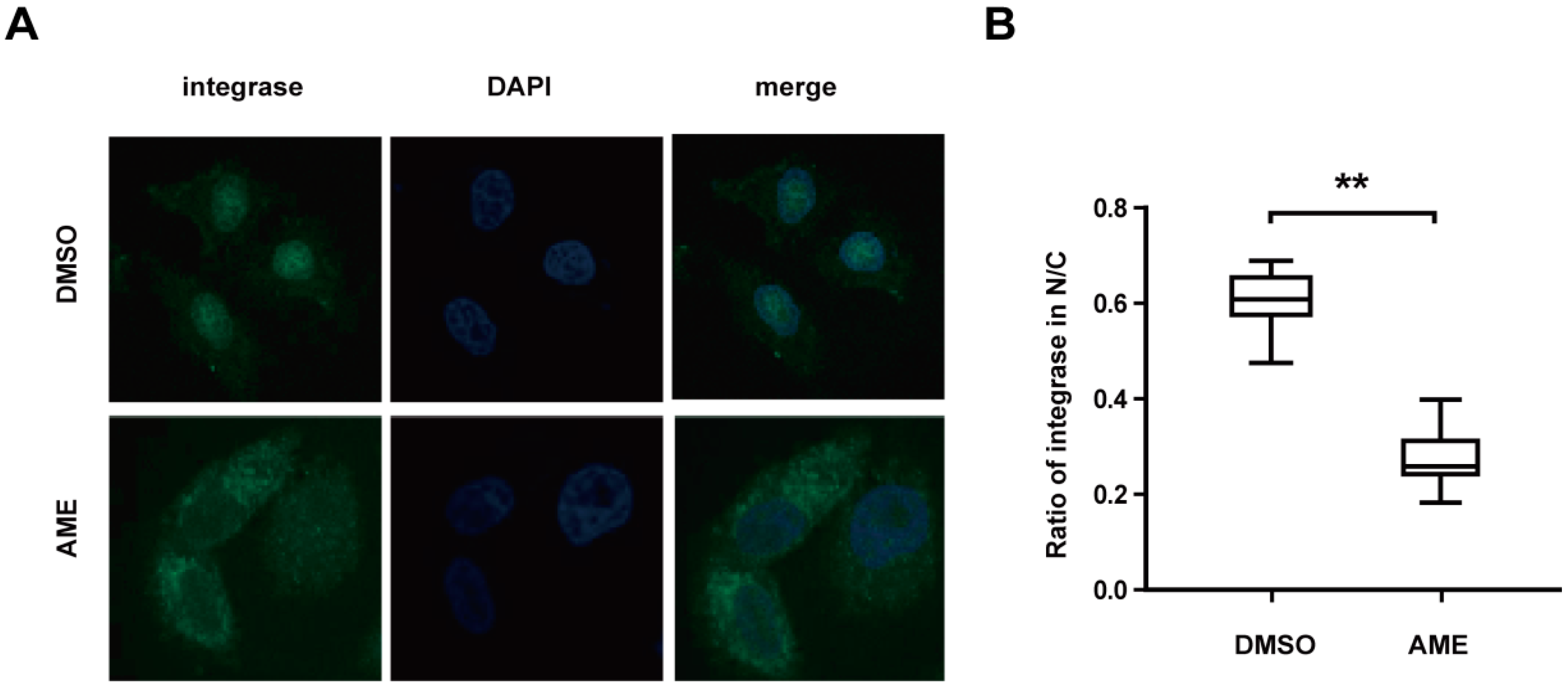
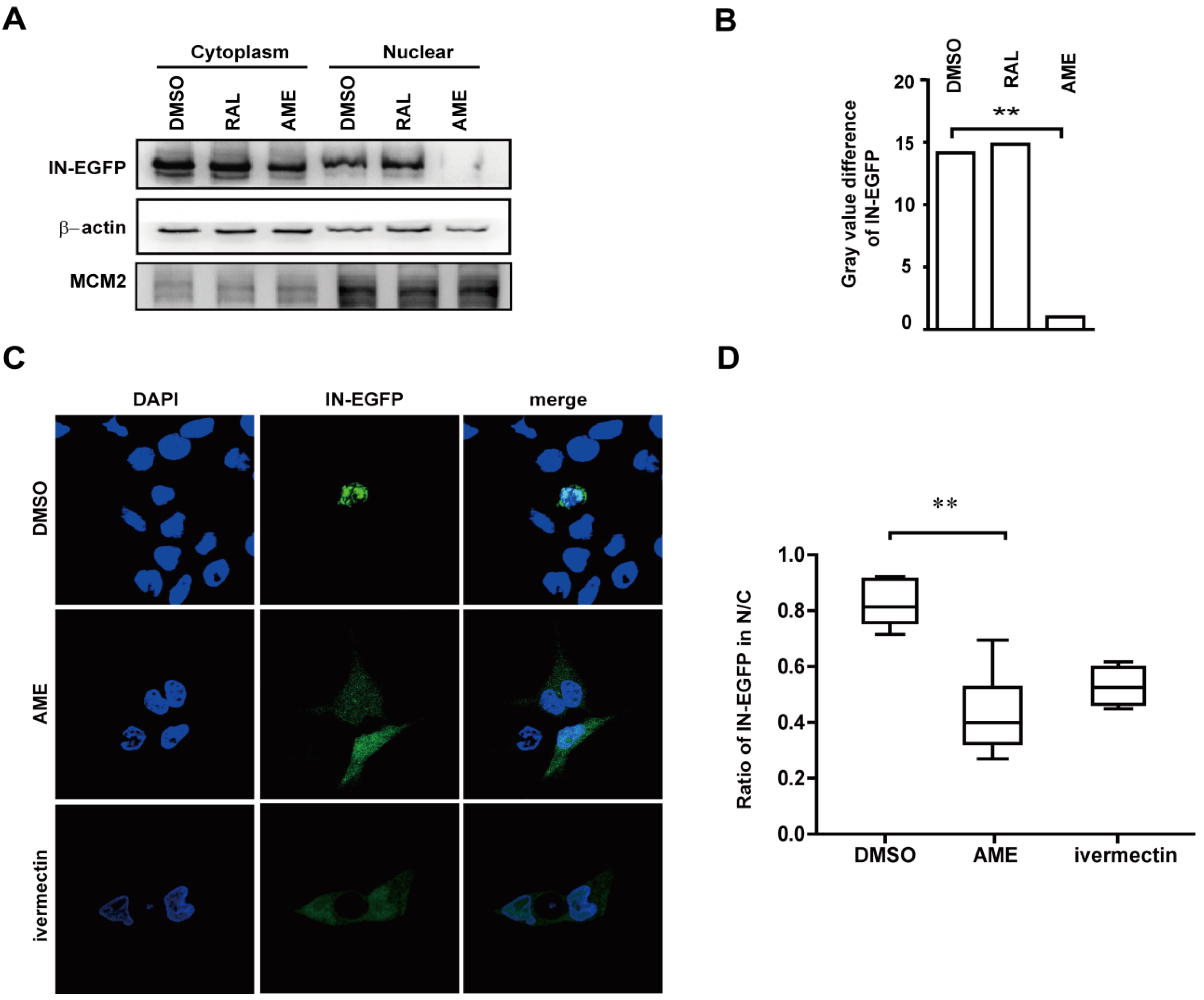
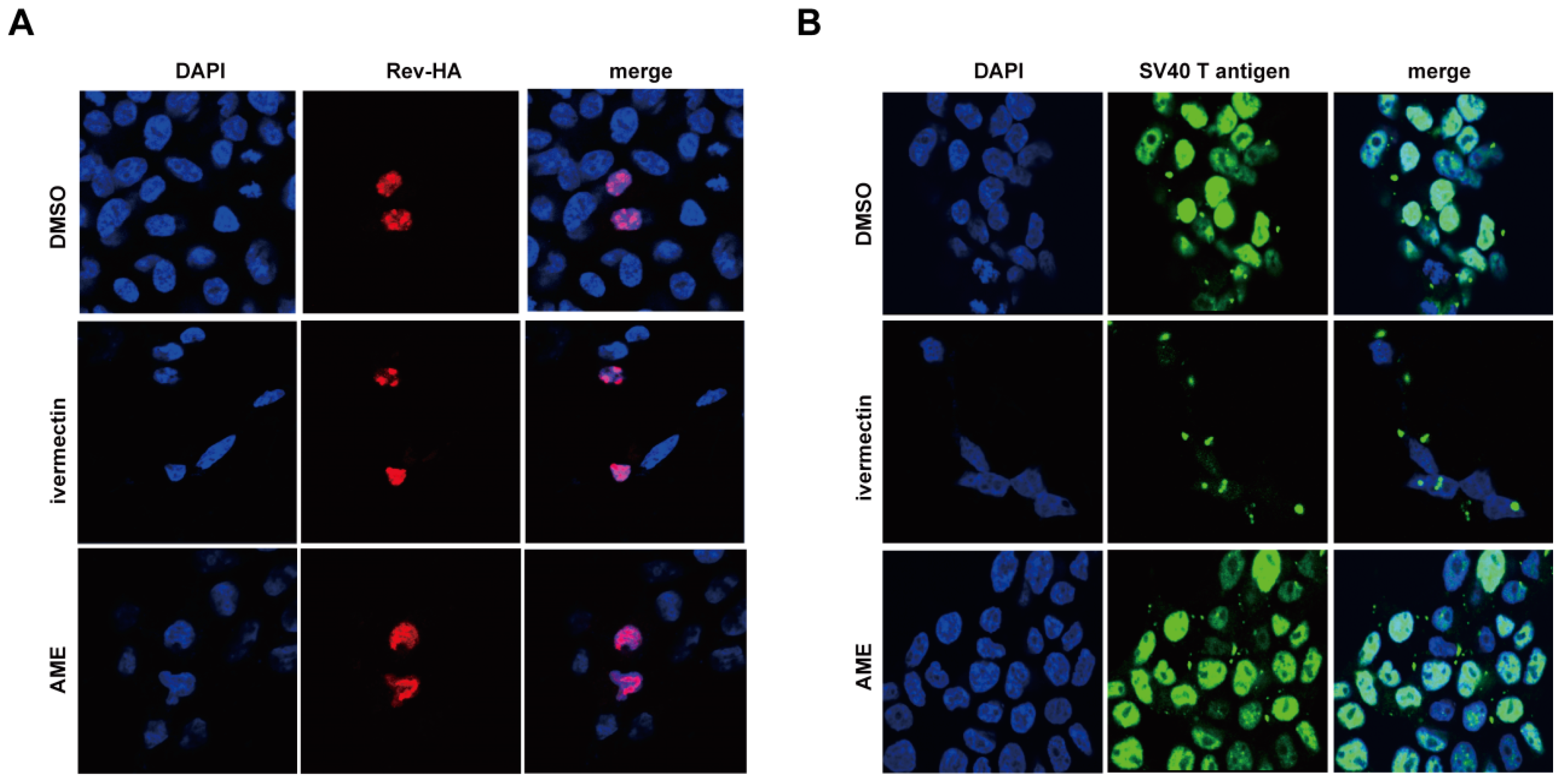
3.3. AME Blocks Nuclear Transport of PIC
3.4. AME Inhibits the Infection of Different HIV-1 Strains
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhao, G.; Wang, C.; Liu, C.; Lou, H. New developments in diketo-containing inhibitors of HIV-1 integrase. Mini Rev. Med. Chem. 2007, 7, 707–725. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Zhang, X.; Pais, G.C.; Cowansage, K.; Neamati, N.; Burke, T.R., Jr.; Pommier, Y. Structural determinants for HIV-1 integrase inhibition by beta-diketo acids. J. Biol. Chem. 2002, 277, 12596–12603. [Google Scholar] [CrossRef] [PubMed]
- McColl, D.J.; Chen, X. Strand transfer inhibitors of HIV-1 integrase: Bringing IN a new era of antiretroviral therapy. Antivir. Res. 2010, 85, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Geretti, A.M.; Armenia, D.; Ceccherini-Silberstein, F. Emerging patterns and implications of HIV-1 integrase inhibitor resistance. Curr. Opin. Infect. Dis. 2012, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.A.; Mesplede, T.; Quashie, P.K. The development of novel HIV integrase inhibitors and the problem of drug resistance. Curr. Opin. Virol. 2012, 2, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Kessl, J.J.; Kvaratskhelia, M. Allosteric inhibition of HIV-1 integrase activity. Curr. Opin. Chem. Biol. 2013, 17, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Depienne, C.; Mousnier, A.; Leh, H.; Le Rouzic, E.; Dormont, D.; Benichou, S.; Dargemont, C. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 2001, 276, 18102–18107. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.P.; Greene, W.C. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 2002, 4, 67–73. [Google Scholar] [CrossRef]
- Raghavendra, N.K.; Shkriabai, N.; Graham, R.; Hess, S.; Kvaratskhelia, M.; Wu, L. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology 2010, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Luban, J. HIV-1 infection: Going nuclear with TNPO3/Transportin-SR2 and integrase. Curr. Biol. 2008, 18, R710–R713. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Hayouka, Z.; Friedler, A.; Loyter, A. Transportin 3 and importin alpha are required for effective nuclear import of HIV-1 integrase in virus-infected cells. Nucleus 2010, 1, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Hearps, A.C.; Jans, D.A. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 2006, 398, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Ao, Z.; Danappa Jayappa, K.; Wang, B.; Zheng, Y.; Kung, S.; Rassart, E.; Depping, R.; Kohler, M.; Cohen, E.A.; Yao, X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 2010, 84, 8650–8663. [Google Scholar] [CrossRef] [PubMed]
- Bouyac-Bertoia, M.; Dvorin, J.D.; Fouchier, R.A.; Jenkins, Y.; Meyer, B.E.; Wu, L.I.; Emerman, M.; Malim, M.H. HIV-1 infection requires a functional integrase NLS. Mol. Cell. 2001, 7, 1025–1035. [Google Scholar] [CrossRef]
- Levin, A.; Armon-Omer, A.; Rosenbluh, J.; Melamed-Book, N.; Graessmann, A.; Waigmann, E.; Loyter, A. Inhibition of HIV-1 integrase nuclear import and replication by a peptide bearing integrase putative nuclear localization signal. Retrovirology 2009, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Bukrinsky, M.I.; Haggerty, S.; Dempsey, M.P.; Sharova, N.; Adzhubei, A.; Spitz, L.; Lewis, P.; Goldfarb, D.; Emerman, M.; Stevenson, M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 1993, 365, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Ambrose, Z.; Martin, T.D.; Oztop, I.; Mulky, A.; Julias, J.G.; Vandegraaff, N.; Baumann, J.G.; Wang, R.; Yuen, W.; et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Haffar, O.K.; Popov, S.; Dubrovsky, L.; Agostini, I.; Tang, H.; Pushkarsky, T.; Nadler, S.G.; Bukrinsky, M. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 2000, 299, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Matreyek, K.A.; Engelman, A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 2011, 85, 7818–7827. [Google Scholar] [CrossRef] [PubMed]
- Monette, A.; Pante, N.; Mouland, A.J. Examining the requirements for nucleoporins by HIV-1. Future Microbiol. 2011, 6, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Christ, F.; Thys, W.; de Rijck, J.; Gijsbers, R.; Albanese, A.; Arosio, D.; Emiliani, S.; Rain, J.C.; Benarous, R.; Cereseto, A.; et al. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 2008, 18, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, L.; Cherepanov, P.; Leyens, L.; Wilson, S.J.; Rasaiyaah, J.; Fassati, A. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology 2009, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Ao, Z.; Huang, G.; Yao, H.; Xu, Z.; Labine, M.; Cochrane, A.W.; Yao, X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 2007, 282, 13456–13467. [Google Scholar] [CrossRef] [PubMed]
- Mousnier, A.; Leh, H.; Mouscadet, J.F.; Dargemont, C. Nuclear import of HIV-1 integrase is inhibited in vitro by styrylquinoline derivatives. Mol. Pharmacol. 2004, 66, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Rawlinson, S.M.; Hearps, A.C.; Jans, D.A. An AlphaScreen(R)-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen 2011, 16, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Marsden, M.D.; Avancena, P.; Kitchen, C.M.; Hubbard, T.; Zack, J.A. Single mutations in HIV integrase confer high-level resistance to raltegravir in primary human macrophages. Antimicrob. Agents Chemother. 2011, 55, 3696–3702. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Otsuka, N.; Matsuo, K.; Shiino, T.; Kojima, A.; Kurata, T.; Sakai, K.; Yamamoto, N.; Isomura, S.; Dhole, T.N.; et al. An infectious DNA clone of HIV type 1 subtype C. AIDS Res. Hum. Retrovir. 1999, 15, 1321–1324. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Gonzalez, J.F.; Salazar, M.G.; Keele, B.F.; Learn, G.H.; Giorgi, E.E.; Li, H.; Decker, J.M.; Wang, S.; Baalwa, J.; Kraus, M.H.; et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 2009, 206, 1273–1289. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ding, J.; Ding, K.; Chen, D.; Cen, S.; Ge, M. Phomonaphthalenone A: A novel dihydronaphthalenone with anti-HIV activity from Phomopsis sp. HCCB04730. Phytochem. Lett. 2013, 6, 257–260. [Google Scholar] [CrossRef]
- Desimmie, B.A.; Schrijvers, R.; Demeulemeester, J.; Borrenberghs, D.; Weydert, C.; Thys, W.; Vets, S.; van Remoortel, B.; Hofkens, J.; de Rijck, J.; et al. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013, 10, 57. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.; Zhao, J.; Yang, Z.; Ma, L.; Mi, Z.; Wu, Y.; Guo, J.; Zhou, J.; Li, X.; Guo, Y.; et al. Microbial Natural Product Alternariol 5-O-Methyl Ether Inhibits HIV-1 Integration by Blocking Nuclear Import of the Pre-Integration Complex. Viruses 2017, 9, 105. https://doi.org/10.3390/v9050105
Ding J, Zhao J, Yang Z, Ma L, Mi Z, Wu Y, Guo J, Zhou J, Li X, Guo Y, et al. Microbial Natural Product Alternariol 5-O-Methyl Ether Inhibits HIV-1 Integration by Blocking Nuclear Import of the Pre-Integration Complex. Viruses. 2017; 9(5):105. https://doi.org/10.3390/v9050105
Chicago/Turabian StyleDing, Jiwei, Jianyuan Zhao, Zhijun Yang, Ling Ma, Zeyun Mi, Yanbing Wu, Jiamei Guo, Jinmin Zhou, Xiaoyu Li, Ying Guo, and et al. 2017. "Microbial Natural Product Alternariol 5-O-Methyl Ether Inhibits HIV-1 Integration by Blocking Nuclear Import of the Pre-Integration Complex" Viruses 9, no. 5: 105. https://doi.org/10.3390/v9050105





