HBV Drug Resistance Substitutions Existed before the Clinical Approval of Nucleos(t)ide Analogues: A Bioinformatic Analysis by GenBank Data Mining
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Search and Qualification Strategy
2.2. Data Extraction
2.3. HBV Genotyping and Detection of Recombinant Sequences
2.4. Systematic Analysis of Potential NUCr Substitutions
2.5. HBV Drug Resistance Substitution Analysis
2.6. Statistical Analysis
3. Results
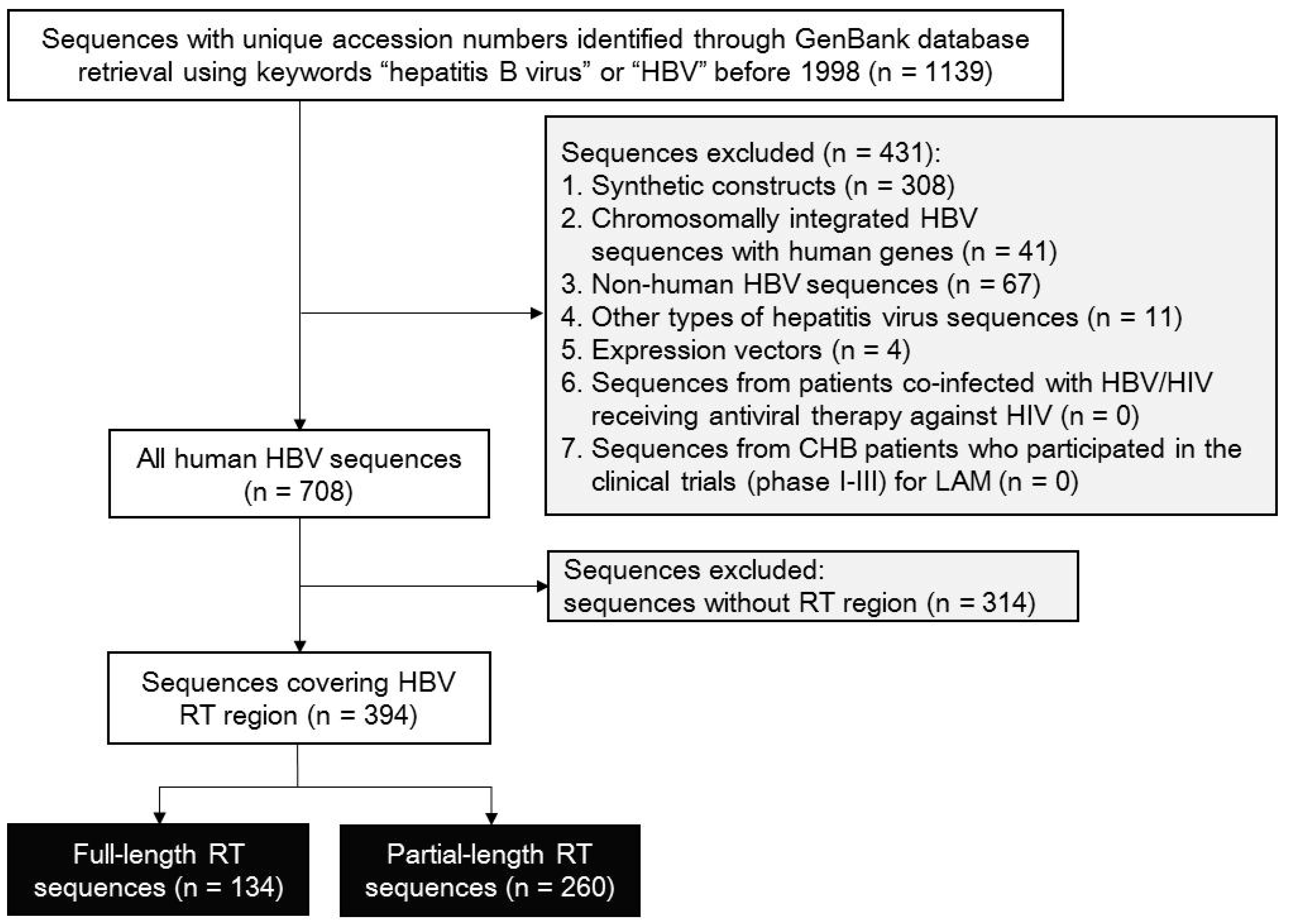
3.1. Selection for RT-Containing HBV Sequences
3.2. Quality Control for HBV RT-Containing Sequences
3.3. RT Sequence Origins and HBV Genotypes
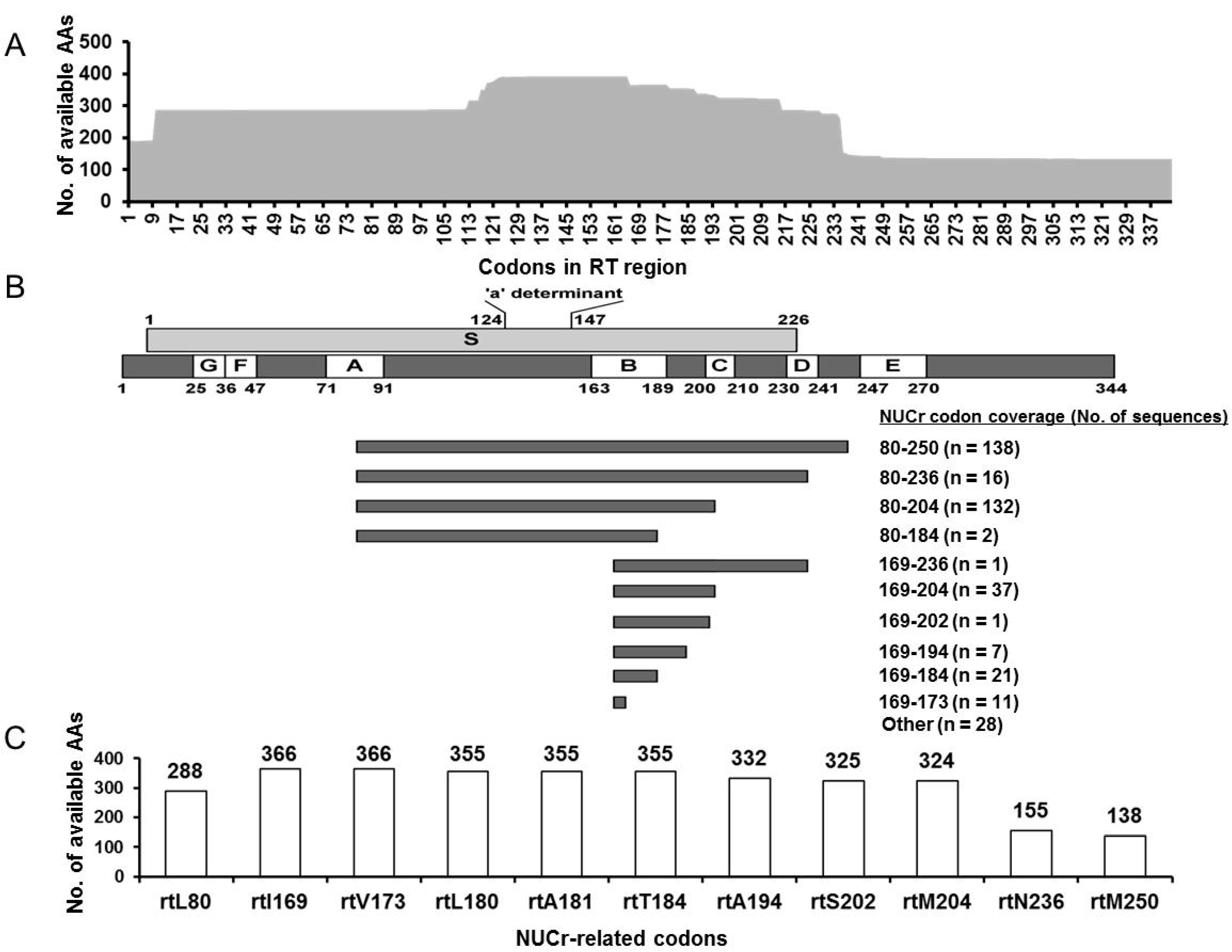
3.4. NUCr Substitution Analysis
3.5. Characterization of the RT Sequences Harboring AA Substitutions Associated with NUCr
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- Zoulim, F.; Locarnini, S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009, 137, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Ferir, G.; Kaptein, S.; Neyts, J. Antiviral treatment of chronic hepatitis B virus (HBV) infections. Viruses 2010, 2, 1279–1305. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Arias, L.; Alvarez, M.; Pacheco, B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: Mechanism of action and resistance. Curr. Opin. Virol. 2014, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hepatol, J. European association for the study of the liver. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–397. [Google Scholar] [CrossRef]
- Girones, R.; Miller, R.H. Mutation rate of the hepadnavirus genome. Virology 1989, 170, 595–597. [Google Scholar] [CrossRef]
- Kang, L.; Pan, J.; Wu, J.; Hu, J.; Sun, Q.; Tang, J. Anti-HBV drugs: Progress, unmet needs, and new hope. Viruses 2015, 7, 4960–4977. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Zhao, P.; Wang, Y.; Chen, L.; Xin, S.; Zhang, X.X.; Xu, D. Investigation into drug-resistant mutations of HBV from 845 nucleoside/nucleotide analogue-naïve Chinese patients with chronic HBV infection. Antivir. Ther. 2015, 20, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Huang, L.H.; Liu, C.M.; Yang, S.; Li, J.; Lin, Z.M.; Kong, X.F.; Yu, D.M.; Zhang, D.H.; Jin, G.D.; et al. Characterization of hepatitis B virus reverse transcriptase sequences in Chinese treatment naïve patients. J. Gastroenterol. Hepatol. 2009, 24, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.W.; Ge, G.H.; Zhao, W.; Gan, J.H.; Zhao, Y.; Niu, Z.L.; Zhang, D.J.; Chen, L.; Yu, X.J.; Yang, L.J. YMDD motif mutations in chronic hepatitis B antiviral treatment naïve patients: A multi-center study. Braz. J. Infect. Dis. 2012, 16, 250–255. [Google Scholar] [CrossRef]
- Liu, B.M.; Li, T.; Xu, J.; Li, X.G.; Dong, J.P.; Yan, P.; Yang, J.X.; Yan, L.; Gao, Z.Y.; Li, W.P.; et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naïve Chinese patients. Antivir. Res. 2010, 85, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Terrault, N.A.; Bzowej, N.H.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Murad, M.H. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016, 63, 261–283. [Google Scholar] [CrossRef] [PubMed]
- Sarin, S.K.; Kumar, M.; Lau, G.K.; Abbas, Z.; Chan, H.L.; Chen, C.J.; Chen, D.S.; Chen, H.L.; Chen, P.J.; Chien, R.N.; et al. Asian-pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.; Bartholomeusz, A.; Locarnini, S. HBV drug resistance: Mechanisms, detection and interpretation. J. Hepatol. 2006, 44, 593–606. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for the Prevention, Care and Treatment of Persons with Chronic Hepatitis B Infection; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Lok, A.S.; Zoulim, F.; Locarnini, S.; Bartholomeusz, A.; Ghany, M.G.; Pawlotsky, J.M.; Liaw, Y.F.; Mizokami, M.; Kuiken, C. Antiviral drug-resistant HBV: Standardization of nomenclature and assays and recommendations for management. Hepatology 2007, 46, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Murakami, S.; Omagari, K.; Matsui, T.; Iio, E.; Isogawa, M.; Watanabe, T.; Karino, Y.; Tanaka, Y. Characterization of novel entecavir resistance mutations. J. Hepatol. 2015, 63, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Shirvani-Dastgerdi, E.; Winer, B.Y.; Celia-Terrassa, T.; Kang, Y.; Tabernero, D.; Yagmur, E.; Rodriguez-Frias, F.; Gregori, J.; Luedde, T.; Trautwein, C.; et al. Selection of the highly replicative and partially multidrug resistant rtS78T HBV polymerase mutation during TDF-ETV combination therapy. J. Hepatol. 2017, 67, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.H.; Garcia, R.T.; Trinh, H.N.; Nguyen, H.A.; Nguyen, K.K.; Nguyen, L.H.; Levitt, B. Prevalence of hepatitis B virus DNA polymerase mutations in treatment-naïve patients with chronic hepatitis B. Aliment. Pharmacol. Ther. 2009, 30, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Vutien, P.; Trinh, H.N.; Garcia, R.T.; Nguyen, H.A.; Levitt, B.S.; Nguyen, K.; da Silveira, E.; Daugherty, T.; Ahmed, A.; Garcia, G.; et al. Mutations in HBV DNA polymerase associated with nucleos(t)ide resistance are rare in treatment-naïve patients. Clin. Gastroenterol. Hepatol. 2014, 12, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Isgro, G.; Di Stefano, R.; Ferraro, D.; Maimone, S.; Brancatelli, S.; Squadrito, G.; Di Marco, V.; Craxi, A.; Raimondo, G. Variability of reverse transcriptase and overlapping S gene in hepatitis B virus isolates from untreated and lamivudine-resistant chronic hepatitis B patients. Antivir. Ther. 2009, 14, 649–654. [Google Scholar] [PubMed]
- Nishijima, N.; Marusawa, H.; Ueda, Y.; Takahashi, K.; Nasu, A.; Osaki, Y.; Kou, T.; Yazumi, S.; Fujiwara, T.; Tsuchiya, S.; et al. Dynamics of hepatitis B virus quasispecies in association with nucleos(t)ide analogue treatment determined by ultra-deep sequencing. PLoS ONE 2012, 7, e35052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes-Gouvea, M.S.; Ferreira, A.C.; Teixeira, R.; Andrade, J.R.; Ferreira, A.S.; Barros, L.M.; Rezende, R.E.; Nastri, A.C.; Leite, A.G.; Piccoli, L.Z.; et al. HBV carrying drug-resistance mutations in chronically infected treatment-naïve patients. Antivir. Ther. 2015, 20, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liao, Y.; Cai, B.; Li, Y.; Li, L.; Zhang, J.; An, Y.; Wang, L. Incidence of natural resistance mutations in naïve chronic hepatitis B patients: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2015, 30, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Zoulim, F. Hepatitis B virus resistance to entecavir in nucleoside naïve patients: Does it exist? Hepatology 2006, 44, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.M.; Huang, Q.W.; Qin, Y.Q.; He, Y.Z.; Qin, H.J.; Zhou, Y.N.; Xu, X.; Huang, M.J. YMDD mutations in patients with chronic hepatitis B untreated with antiviral medicines. World J. Gastroenterol. 2005, 11, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Karatayli, E.; Karayalcin, S.; Karaaslan, H.; Kayhan, H.; Turkyilmaz, A.R.; Sahin, F.; Yurdaydin, C.; Bozdayi, A.M. A novel mutation pattern emerging during lamivudine treatment shows cross-resistance to adefovir dipivoxil treatment. Antivir. Ther. 2007, 12, 761–768. [Google Scholar] [PubMed]
- Liu, Y.; Xu, Z.H.; Wang, Y.; Li, X.D.; Liu, L.M.; Chen, L.; Xin, S.J.; Xu, D.P. rtM204Q may serve as a novel lamivudine-resistance-associated mutation of hepatitis B virus. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Bozdayi, A.M.; Uzunalimoglu, O.; Turkyilmaz, A.R.; Aslan, N.; Sezgin, O.; Sahin, T.; Bozdayi, G.; Cinar, K.; Pai, S.B.; Pai, R.; et al. YSDD: A novel mutation in HBV DNA polymerase confers clinical resistance to lamivudine. J. Viral Hepat. 2003, 10, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Xiang, K.H.; Zhao, C.Y.; Wang, S.; Li, Y.; Su, M.Z.; Wang, Q.Y.; Xu, X.Z.; Deng, J.; Zhuang, H.; Li, T. Impacts of HBV rtH55R polymerase substitution on viral replication and rtM204I/V resistance to nucleoside/nucleotide antiviral drugs. Antivir. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Chien, R.N.; Hu, C.C.; Lai, M.W.; Yeh, C.T. Identification of hepatitis B virus rtS117F substitution as a compensatory mutation for rtM204I during lamivudine therapy. J. Antimicrob. Chemother. 2012, 67, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Torresi, J.; Earnest-Silveira, L.; Civitico, G.; Walters, T.E.; Lewin, S.R.; Fyfe, J.; Locarnini, S.A.; Manns, M.; Trautwein, C.; Bock, T.C. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology 2002, 299, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.B.; Bozdayi, A.M.; Pai, R.B.; Beker, T.; Sarioglu, M.; Turkyilmaz, A.R.; Grier, J.; Yurdaydin, C.; SChinazi, R.F. Emergence of a novel mutation in the FLLA region of hepatitis B virus during lamivudine therapy. Antimicrob. Agents Chemother. 2005, 49, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Cheng, Y.C. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(-)SddC (3TC) resistance. Biochem. Pharmacol. 1998, 55, 1567–1572. [Google Scholar] [CrossRef]
- Zollner, B.; Sterneck, M.; Wursthorn, K.; Petersen, J.; Schroter, M.; Laufs, R.; Feucht, H.H. Prevalence, incidence, and clinical relevance of the reverse transcriptase V207I mutation outside the YMDD motif of the hepatitis B virus polymerase during lamivudine therapy. J. Clin. Microbiol. 2005, 43, 2503–2505. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Liu, Y.; Si, L.L.; Li, L.; Chen, G.F.; Xin, S.J.; Zhao, J.M.; Xu, D. Variable influence of mutational patterns in reverse-transcriptase domain on replication capacity of hepatitis B virus isolates from antiviral-experienced patients. Clin. Chim. Acta 2011, 412, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Liu, Y.; Li, L.; Xu, Z.H.; Si, L.L.; Dai, J.Z.; Li, X.D.; Wang, L.; Yao, Z.T.; Xin, S.J.; et al. The rtL229 substitutions in the reverse transcriptase region of hepatitis B virus (HBV) polymerase are potentially associated with lamivudine resistance as a compensatory mutation. J. Clin. Virol. 2012, 54, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Zhang, B.; Zhang, X.D.; He, T.T.; Xu, W.Y.; Fu, L.J.; Tu, C.Y. Substitution rtQ267H of hepatitis B virus increases the weight of replication and lamivudine resistance. Hepat. Mon. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Kim, D.H.; Lee, A.R.; Kim, B.K.; Park, Y.K.; Park, E.S.; Ahn, S.H.; Shin, G.C.; Park, S.; Kang, H.S.; et al. Substitution at rt269 in hepatitis B virus polymerase is a compensatory mutation associated with multi-drug resistance. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Cento, V.; Van Hemert, F.; Neumann-Fraune, M.; Mirabelli, C.; Di Maio, V.C.; Salpini, R.; Bertoli, A.; Micheli, V.; Gubertini, G.; Romano, S.; et al. Anti-HBV treatment induces novel reverse transcriptase mutations with reflective effect on HBV S antigen. J. Infect. 2013, 67, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, X.; Xin, S.; Xu, Z.; Chen, R.; Yang, J.; Liu, L.; Wong, V.W.; Yang, D.; Chan, H.L.; et al. The rtA181S mutation of hepatitis B virus primarily confers resistance to adefovir dipivoxil. J. Viral Hepat. 2015, 22, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Wang, J.H.; Du, S.C.; Tian, J.H.; Yang, R.F.; Wei, L. rtE218G, a novel hepatitis B virus mutation with resistance to adefovir dipivoxil in patients with chronic hepatitis B. J. Viral Hepat. 2010, 17, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Schildgen, O.; Olotu, C.; Funk, A.; Zollner, B.; Helm, M.; Rockstroh, J.K.; Sirma, H. Selection and counterselection of the rtI233V adefovir resistance mutation during antiviral therapy. J. Clin. Microbiol. 2010, 48, 631–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, W.H.; Li, X.D.; Xu, Z.H.; Wang, X.; Li, C.X.; Chen, L.; Xin, S.J.; Xu, D.P. Screening and identification of a novel adefovir dipivoxil resistance associated mutation, rtN236V, of HBV from a large cohort of HBV-infected patients. Antivir. Ther. 2014, 19, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Warner, N.; Locarnini, S.; Kuiper, M.; Bartholomeusz, A.; Ayres, A.; Yuen, L.; Shaw, T. The L80I substitution in the reverse transcriptase domain of the hepatitis B virus polymerase is associated with lamivudine resistance and enhanced viral replication in vitro. Antimicrob. Agents Chemother. 2007, 51, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.; Rodes, B.; Zoulim, F.; Bartholomeusz, A.; Soriano, V. Mutations affecting the replication capacity of the hepatitis B virus. J. Viral Hepat. 2006, 13, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Ghany, M.; Liang, T.J. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology 2007, 132, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Bottecchia, M.; Madejon, A.; Sheldon, J.; Garcia-Samaniego, J.; Barreiro, P.; Soriano, V. Hepatitis B virus genotype A2 harbours an L217R polymorphism which may account for a lower response to adefovir. J. Antimicrob. Chemother. 2008, 62, 626–627. [Google Scholar] [CrossRef] [PubMed]
- Karatayli, E.; Karatayli, S.C.; Cinar, K.; Gokahmetoglu, S.; Guven, K.; Idilman, R.; Yurdaydin, C.; Bozdayi, A.M. Molecular characterization of a novel entecavir mutation pattern isolated from a multi-drug refractory patient with chronic hepatitis B infection. J. Clin. Virol. 2012, 53, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Cui, J.; Guo, J.J.; Zhang, W.L.; Cai, X.F.; Yuan, Z.W.; Li, Q.L.; Deng, X.Y.; Zeng, A.Z.; Hu, Y.; et al. Phenotypic assay of a hepatitis B virus strain carrying an rtS246T variant using a new strategy. J. Med. Virol. 2012, 84, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Budeus, B.; Cao, L.; Wu, C.C.; Wang, Y.; Zhang, X.Y.; Rayner, S.; Hoffmann, D.; Lu, M.J.; Chen, X.W. The amino acid substitutions rtP177G and rtF249A in the reverse transcriptase domain of hepatitis B virus polymerase reduce the susceptibility to tenofovir. Antivir. Res. 2013, 97, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.; Clark, C.; Khan, A.; Kellam, P.; Tedder, R. Genotyping hepatitis B virus from whole- and sub-genomic fragments using position-specific scoring matrices in HBV STAR. J. Gen. Virol. 2006, 87, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.K.; Bulla, I.; Abdou-Chekaraou, M.; Gordien, E.; Morgenstern, B.; Zoaulim, F.; Deny, P.; Stanke, M. jpHMM: Recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res. 2012, 40, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiang, K.; Li, Y.; Li, Y.; Deng, J.; Xu, X.; Yan, L.; Zhuang, H.; Li, T. Higher detection rates of amino acid substitutions in HBV reverse transcriptase/surface protein overlapping sequence is correlated with lower serum HBV DNA and HBsAg levels in HBeAg-positive chronic hepatitis B patients with subgenotype B2. Infect. Genet. Evol. 2016, 40, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Liu, B.M.; Li, X.G.; Yan, C.H.; Xu, J.; Sun, X.W.; Wang, Y.H.; Jiao, X.J.; Yan, L.; Dong, J.P.; et al. Profile of HBV antiviral resistance mutations with distinct evolutionary pathways against nucleoside/nucleotide analogue treatment among Chinese chronic hepatitis B patients. Antivir. Ther. 2010, 15, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Zoulim, F.; Locarnini, S. Optimal management of chronic hepatitis B patients with treatment failure and antiviral drug resistance. Liver Int. 2013, 33, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J. Gastroenterol. 2014, 20, 5427–5434. [Google Scholar] [CrossRef] [PubMed]
- Fujiyama, A.; Miyanohara, A.; Nozaki, C.; Yoneyama, T.; Ohtomo, N.; Matsubara, K. Cloning and structural analyses of hepatitis B virus DNAs, subtype adr. Nucleic Acids Res. 1983, 11, 4601–4610. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, H.; Inokoshi, J.; Namiki, M.; Shimada, J.; Omura, S. Structural analysis of the gene coding for hepatitis B virus surface antigen and its product. J. Gen. Virol. 1985, 66 Pt 1, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Mukaide, M.; Kumazawa, T.; Hoshi, A.; Kawaguchi, R.; Hikiji, K. The complete nucleotide sequence of hepatitis B virus, subtype adr (SRADR) and phylogenetic analysis. Nucleic Acids Res. 1992, 20, 6105. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Omi, S.; Wang, Y.; Itoh, Y.; Tsuda, F.; Tanaka, T.; Akahane, Y.; Miyakawa, Y.; Mayumi, M. The loss of subtypic determinants in alleles, d/y or w/r, on hepatitis B surface antigen. Mol. Immunol. 1989, 26, 197–205. [Google Scholar] [PubMed]
- Rho, H.M.; Kim, K.; Hyun, S.W.; Kim, Y.S. The nucleotide sequence and reading frames of a mutant hepatitis B virus subtype adr. Nucleic Acids Res. 1989, 17, 2124. [Google Scholar] [CrossRef] [PubMed]
- Wands, J.R.; Liang, T.J.; Blum, H.E.; Shafritz, D.A. Molecular pathogenesis of liver disease during persistent hepatitis B virus infection. Semin. Liver Dis. 1992, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, K.; Khandekar, P. Identification and characterization of mutations in X region of a hepatitis B virus carrier. Indian J. Biochem. Biophys. 1992, 29, 494–497. [Google Scholar] [PubMed]
- Karayiannis, P.; Alexopoulou, A.; Hadziyannis, S.; Thursz, M.; Watts, R.; Seito, S.; Thomas, H.C. Fulminant hepatitis associated with hepatitis B virus e antigen-negative infection: Importance of host factors. Hepatology 1995, 22, 1628–1634. [Google Scholar] [PubMed]
- Asahina, Y.; Enomoto, N.; Ogura, Y.; Kurosaki, M.; Sakuma, I.; Izumi, N.; Marumo, F.; Sato, C. Sequential changes in full-length genomes of hepatitis B virus accompanying acute exacerbation of chronic hepatitis B. J. Hepatol. 1996, 25, 787–794. [Google Scholar] [CrossRef]
- Fang, D.G.R.; Zhang, Q.; Li, Z.; Duan, S.; Yin, Z. Transient expression and antigenic characterization of HBsAg 126Ser of hepatitis B virus aa126Ile->Ser mutant. Chin. J. Virol. 1998, 14, 1–9. [Google Scholar]
- Bowyer, S.M.; van Staden, L.; Kew, M.C.; Sim, J.G. A unique segment of the hepatitis B virus group a genotype identified in isolates from South Africa. J. Gen. Virol. 1997, 78, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Akahane, Y.; Hino, K.; Ohta, Y.; Mishiro, S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: Comparative analysis of 40 full-length isolates. Arch. Virol. 1998, 143, 2313–2326. [Google Scholar] [CrossRef] [PubMed]
- Grethe, S.; Monazahian, M.; Bohme, I.; Thomssen, R. Characterization of unusual escape variants of hepatitis B virus isolated from a hepatitis B surface antigen-negative subject. J. Virol. 1998, 72, 7692–7696. [Google Scholar] [PubMed]
- Mbayed, V.A.; Lopez, J.L.; Telenta, P.F.S.; Palacios, G.; Badia, I.; Ferro, A.; Galoppo, C.; Campos, R. Distribution of hepatitis B virus genotypes in two different pediatric populations from Argentina. J. Clin. Microbiol. 1998, 36, 3362–3365. [Google Scholar] [CrossRef] [PubMed]
- Poovorawan, Y.; Sripattanawat, R.; Theamboonlers, A.; Chongsrisawat, V.; Nuchprayoon, I. Hepatocellular carcinoma: Significance of HBV vertical transmission. Asian Pac. J. Allergy Immunol. 1998, 16, 93–103. [Google Scholar] [PubMed]
- Colonno, R.J.; Rose, R.; Baldick, C.J.; Levine, S.; Pokornowski, K.; Yu, C.F.; Walsh, A.; Fang, J.; Hsu, M.; Mazzucco, C.; et al. Entecavir resistance is rare in nucleoside naïve patients with hepatitis B. Hepatology 2006, 44, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Tenney, D.J.; Levine, S.M.; Rose, R.E.; Walsh, A.W.; Weinheimer, S.P.; Discotto, L.; Plym, M.; Pokornowski, K.; Yu, C.F.; Angus, P.; et al. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to lamivudine. Antimicrob. Agents Chemother. 2004, 48, 3498–3507. [Google Scholar] [CrossRef] [PubMed]
- Langley, D.R.; Walsh, A.W.; Baldick, C.J.; Eggers, B.J.; Rose, R.E.; Levine, S.M.; Kapur, A.J.; Colonno, R.J.; Tenney, D.J. Inhibition of hepatitis B virus polymerase by entecavir. J. Virol. 2007, 81, 3992–4001. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, A.; Locarnini, S. Hepatitis B virus mutations associated with antiviral therapy. J. Med. Virol. 2006, 78, S52–S55. [Google Scholar] [CrossRef] [PubMed]
- Baldick, C.J.; Tenney, D.J.; Mazzucco, C.E.; Eggers, B.J.; Rose, R.E.; Pokornowski, K.A.; Yu, C.F.; Colonno, R.J. Comprehensive evaluation of hepatitis B virus reverse transcriptase substitutions associated with entecavir resistance. Hepatology 2008, 47, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Widasari, D.I.; Yano, Y.; Heriyanto, D.S.; Utsumi, T.; Yamani, L.N.; Rinonce, H.T.; Wasityastuti, W.; Lusida, M.I.; Soetjipto; Okada, R.; et al. A deep-sequencing method detects drug-resistant mutations in the hepatitis B virus in Indonesians. Intervirology 2014, 57, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Chang, H.Y.; Lim, S.M.; Kim, S.U.; Park, J.Y.; Kim, J.K.; Lee, K.S.; Han, K.H.; Chon, C.Y.; Ahn, S.H. Quasispecies and pre-existing drug-resistant mutations of hepatitis B virus in patients with chronic hepatitis B. Gut Liver 2013, 7, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Jarabo, C.M.; Arias, A.; Baranowski, E.; Escarmis, C.; Domingo, E. Memory in viral quasispecies. J. Virol. 2000, 74, 3543–3547. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.; Ruiz-Jarabo, C.M.; Escarmis, C.; Domingo, E. Fitness increase of memory genomes in a viral quasispecies. J. Mol. Biol. 2004, 339, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Arias, L.; Sebastian-Martin, A.; Alvarez, M. Viral reverse transcriptases. Virus Res. 2017, 234, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Ono-Nita, S.K.; Kato, N.; Shiratori, Y.; Masaki, T.; Lan, K.H.; Carrilho, F.J.; Omata, M. YMDD motif in hepatitis B virus DNA polymerase influences on replication and lamivudine resistance: A study by in vitro full-length viral DNA transfection. Hepatology 1999, 29, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Margeridon-Thermet, S.; Nguyen, M.H.; Liu, T.F.; Kagan, R.M.; Beggel, B.; Verheyen, J.; Kaiser, R.; Shafer, R.W. Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery. Antivir. Res. 2010, 88, 269–275. [Google Scholar] [CrossRef] [PubMed]
| Drugs | Primary NUCr Substitutions | Secondary Substitutions |
|---|---|---|
| LAM | rtA181T/V [2,4,11], rtA181S [27], rtM204I/V [2,4,11], rtM204Q/S [28,29] | rtH55R [30], rtL80I/V [2,4,11], rtS117F [31], rtT128N [32], rtW153Q [32], rtV173L [2,4,11], rtL180M [2,4,11], rtL180C [33], rtA200V [34], rtV207I [35], rtL217P [36], rtL229F [37], rtQ267H [38], rtL269I [39] |
| ADV | rtS78T [40], rtA181T/V [2,4,11], rtA181S [41], rtE218G [42], rtI233V [43], rtN236T [2,4,11], rtN236V [44] | rtL80I/V [45], rtV84M [46], rtS85A [46], rtV214A [47], rtL217R [48], rtP237H [47] |
| ETV | rtS78T [18], rtI169T [2,4,11], rtA181T/V [2,4,11], rtT184A/C/F/G/I/L/M/S [2,4,11], rtA186T [17], rtS202C/G/I [2,4,11], rtM204I/V [2,4,11], rtM250I/V/L [2,4,11] | rtI163V [17], rtS219A [49], rtY245H [49], rtS246T [50], rtL269I [39] |
| LdT | rtM204I/V [2,4,11], rtA181T/V [2,4,11] | |
| TDF/TAF | rtS78T [18], rtP177G [51], rtA181T/V [2,4,11], rtA194T [2,4,11], rtF249A [51] |
| Substitution Category | NUCr-Related Codons | Substitution Types | No. of Substituted AAs | No. of Available AAs | Substitution Frequency (%) 1 |
|---|---|---|---|---|---|
| Primary | rtI169 | L | 2 | 366 | 0.5 |
| NUCr | rtA181 | G | 1 | 355 | 0.3 |
| substitution | rtT184 | A 3, S 3 | 1, 1 | 355 | 0.6 |
| rtA194 | ND | 0 | 332 | 0 | |
| rtS202 | T, R | 2, 1 | 325 | 0.9 | |
| rtM204 | L | 2 | 324 | 0.6 | |
| rtN236 | ND | 0 | 155 | 0 | |
| rtM250 | K | 1 | 138 | 0.7 | |
| Subtotal | 8 codons | 6 codons, 8 types | 11 | 2350 | 0.5 2 |
| Secondary | rtL80 | V 3 | 7 | 288 | 2.4 |
| substitution | rtV173 | G, A, L 3 | 5, 2, 2 | 366 | 2.5 |
| rtL180 | ND | 0 | 355 | 0 | |
| Subtotal | 3 codons | 2 codons, 4 types | 16 | 1008 | 1.6 2 |
| Total | 11 codons | 8 codons, 12 types | 27 | 3358 | 0.8 |
| Accession Number | Releasing Year | Sample Origin | Sequencing Method | Sequence Length (bp) | RT Coverage (AA) | Genotype | Substitution | References |
|---|---|---|---|---|---|---|---|---|
| X01587.1 | 1983 | Japan | Clone | 3214 | 1–344 | C | V173G | [58] |
| M23808.1 | 1985 | Japan | Clone | 681 | 10–235 | C | V173G | [59] |
| E01164.1 | 1986 | Japan | Clone | 748 | 6–253 | C | V173G | NA |
| M38636.1 | 1988 | South Korea | Clone | 3213 | 1–344 | C | V173G | [60] |
| U19777.1 | 1988 | China | Clone | 731 | 1–243 | C | T184A | NA |
| M27765.1 | 1989 | Japan | Clone | 678 | 10–234 | C | I169L | [61] |
| X14193.1 | 1989 | South Korea | Clone | 3213 | 1–344 | C | V173G | [62] |
| S50225.1 | 1992 | USA | Clone | 3222 | 1–344 | A | V173A | [63] |
| S56208.1 | 1992 | India | Clone | 123 | 169–208 | D | S202T | [64] |
| X77310.1 | 1994 | Italy | Direct | 1401 | 1–300 | D | S202R | NA |
| X80925.1 | 1995 | UK | Direct | 3182 | 1–344 | D | S202T | [65] |
| D50519.1 | 1996 | Japan | Clone | 3215 | 1–344 | C | A181G | [66] |
| AF013631.1 | 1997 | China | Clone | 681 | 10–235 | C | L80V | [67] |
| AF036236.1 | 1997 | China | Clone | 838 | 1–236 | C | L80V | NA |
| AF036237.1 | 1997 | China | Clone | 838 | 1–236 | C | L80V | NA |
| AF036238.1 | 1997 | China | Clone | 838 | 1–236 | C | L80V | NA |
| AF036239.1 | 1997 | China | Clone | 838 | 1–236 | C | L80V | NA |
| U87739.1 | 1997 | South Africa | Direct | 846 | 1–235 | A | L80V | [68] |
| AB014382.1 | 1998 | Japan | Direct | 3215 | 1–344 | C | L80V | [69] |
| AJ003027.1 | 1998 | Germany | Clone | 1158 | 1–235 | D | I169L | [70] |
| AF043564.1 | 1998 | Argentina | Direct | 217 | 122–193 | A | V173A | [71] |
| AJ003026.1 | 1998 | Germany | Clone | 1164 | 1–235 | D | V173L | [70] |
| AJ003028.1 | 1998 | Germany | Clone | 1164 | 1–235 | D | V173L | [70] |
| AF044985.1 | 1998 | Belgium | Clone | 297 | 117–215 | B | T184S | NA |
| AF074449.1 | 1998 | Thailand | Direct | 720 | 10–248 | C | M204L | [72] |
| AF075604.1 | 1998 | Thailand | Direct | 720 | 10–248 | C | M204L | NA |
| AJ012207.1 | 1998 | Germany | Clone | 3221 | 1–344 | A | M250K | NA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Xiang, K.; Su, M.; Li, Y.; Ji, W.; Li, Y.; Zhuang, H.; Li, T. HBV Drug Resistance Substitutions Existed before the Clinical Approval of Nucleos(t)ide Analogues: A Bioinformatic Analysis by GenBank Data Mining. Viruses 2017, 9, 199. https://doi.org/10.3390/v9080199
Xu X, Xiang K, Su M, Li Y, Ji W, Li Y, Zhuang H, Li T. HBV Drug Resistance Substitutions Existed before the Clinical Approval of Nucleos(t)ide Analogues: A Bioinformatic Analysis by GenBank Data Mining. Viruses. 2017; 9(8):199. https://doi.org/10.3390/v9080199
Chicago/Turabian StyleXu, Xizhan, Kuanhui Xiang, Mingze Su, Yao Li, Wei Ji, Yutang Li, Hui Zhuang, and Tong Li. 2017. "HBV Drug Resistance Substitutions Existed before the Clinical Approval of Nucleos(t)ide Analogues: A Bioinformatic Analysis by GenBank Data Mining" Viruses 9, no. 8: 199. https://doi.org/10.3390/v9080199






