Complexation of a Polypeptide-Polyelectrolytes Bioparticle as a Biomaterial of Antibacterial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Bioparticles
2.3. Preparation of Giant Vesicles and Electroporation
2.4. Dynamic Light Scattering and Zeta Potential
2.5. Isothermal Titration Calorimetry (ITC)
2.6. In Vitro Antibacterial Analysis
3. Results and Discussion
3.1. Thermodynamics of Interaction in the Bioparticles Production
3.2. Colloidal Structure
3.3. Bioparticle Interaction with Model Membrane
3.4. Antibacterial Activity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Y.; Liu, Y.; Liu, W.; Lu, C.; Wang, L. Chitosan microparticles ionically cross-linked with poly(γ-glutamic acid) as antimicrobial peptides and nitric oxide delivery systems. Biochem. Eng. J. 2015, 95, 78–85. [Google Scholar] [CrossRef]
- Gao, W.; Chen, Y.; Zhang, Y.; Zhang, Q.; Zhang, L. Nanoparticle-based local antimicrobial drug delivery. Adv. Drug. Deliv. Rev. 2018, 127, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.G.; Hosseinidoust, Z. Liposomes for antibiotic encapsulation and delivery. ACS Infect. Dis. 2020, 6, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fei, X.; Xu, L.; Wang, Y.; Tian, J.; Li, Y. Pressure-sensitive antibacterial hydrogel dressing for wound monitoring in bed ridden patients. J. Colloid Interface Sci. 2022, 627, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Tamahkar, E.; Bakhshpour, M.; Denizil, A. Molecularly imprinted composite bacterial cellulose nanofibers for antibiotic release. J. Biomater. Sci. 2019, 30, 450–461. [Google Scholar] [CrossRef]
- Nair, A.; Mallya, R.; Suvarna, V.; Khan, T.A.; Momin, M.; Omri, A. Nanoparticles—Attractive carriers of antimicrobial essential oils. Antibiotics 2022, 11, 108. [Google Scholar] [CrossRef]
- Boczar, D.; Michalska, K. Cyclodextrin inclusion complexes with antibiotics and antibacterial agents as drug-delivery systems—A pharmaceutical perspective. Pharmaceutics 2022, 14, 1389. [Google Scholar] [CrossRef]
- Catauro, M.; Tranquillo, E.; Poggetto, G.D.; Naviglio, S.; Barrino, F. Antibacterial properties of sol–gel biomaterials with different percentages of PEG or PCL. Macromol. Symp. 2020, 389, 1900056. [Google Scholar] [CrossRef]
- Jian, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug Del. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef]
- Kundu, R. Cationic amphiphilic peptides: Synthetic antimicrobial agents inspired by nature. ChemMedChem 2020, 15, 1887–1896. [Google Scholar] [CrossRef]
- Cirioni, O.; Silvestri, C.; Ghiselli, R.; Kamysz, W.; Minardi, D.; Castelli, P.; Orlando, F.; Kamysz, E.; Provinciali, M.; Muzzonigro, G.; et al. In vitro and in vivo effects of sub-mics of pexiganan and imipenem on Pseudomonas aeruginosa adhesion and biofilm development. Le Infez. Med. 2013, 4, 287–295. [Google Scholar]
- Rajchakit, U.; Sarojini, V. Recent developments in antimicrobial-peptide-conjugated gold nanoparticles. Bioconjugate Chem. 2017, 28, 2673–2686. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for delivery of antimicrobial peptides. Nanomaterials 2020, 10, 560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Angelo, I.; Casciaro, B.; Miro, A.; Quaglia, F.; Mangoni, M.L.; Ungaro, F. Overcoming barriers in Pseudomonas aeruginosa lung infections: Engineered nanoparticles for local delivery of a cationic antimicrobial peptide. Coll. Surf. B Biointerfaces 2015, 135, 717–725. [Google Scholar] [CrossRef]
- Ma, B.; Chen, Y.; Hu, G.; Zeng, Q.; Lv, X.; Hwan Oh, D.; Fu, X.; Jin, Y. Ovotransferrin antibacterial peptide coupling mesoporous silica nanoparticle as an effective antibiotic delivery system for treating bacterial infection in vivo. ACS Biomater. Sci. Eng. 2022, 8, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Sonia, T.A.; Sharma, C.P. Chitosan and its derivatives for drug delivery perspective. Adv. Polym. Sci. 2011, 243, 23–53. [Google Scholar]
- Rodrigues, S.; Dionisio, M.; Lopez, C.R.; Greha, A. Biocompatibility of chitosan carriers with application in drug delivery. J. Funct. Biomater. 2012, 3, 615–641. [Google Scholar] [CrossRef] [Green Version]
- Mulinti, P.; Shreffler, J.; Hasan, R.; Dea, M.; Brooks, A.E. Infection responsive smart delivery of antibiotics using recombinant spider silk nanospheres. Pharmaceutics 2021, 13, 1358. [Google Scholar] [CrossRef]
- Rinaldi, F.; Hanieh, P.; Sennato, S.; De Santis, F.; Forte, J.; Fraziano, M.; Casciardi, S.; Marianecci, C.; Bordi, F.; Carafa, M. Rifampicin–liposomes for Mycobacterium abscessus infection treatment: Intracellular uptake and antibacterial activity evaluation. Pharmaceutics 2021, 13, 1070. [Google Scholar] [CrossRef]
- Pabon, B.Y. Synthesis of Peptide-Functionalized Chitosan Polymeric Nanoparticles and Evaluation of Their In Vitro Activity against Escherichia coli and Staphylococcus aureus. Master’s Thesis, Industrial University of Santander, Faculty of Sciences, Bucaramanga, Colombia, 2015; 33p. [Google Scholar]
- Philibert, T.; Lee, B.H.; Fabien, N. Current status and new perspectives on chitin and chitosan as functional biopolymers. Appl. Biochem. Biotechnol. 2017, 181, 1314–1337. [Google Scholar] [CrossRef]
- Collado-Gonzalez, M.; Esteban, M.A. Chitosan-nanoparticles effects on mucosal immunity: A systematic review. Fish Shellfish Immunol. 2022, 130, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bannikova, A.; Rasumova, L.; Evteev, A.; Evdokimov, I.; Kasapis, S. Protein-loaded sodium alginate and carboxymethyl cellulose beads for controlled release under simulated gastrointestinal conditions. Food Sci. Technol. 2017, 52, 2171–2179. [Google Scholar] [CrossRef]
- Tonnesen, H.H.; Karlsen, J. Alginate in drug delivery systems. Drug Dev. Ind. Pharm. 2002, 28, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Patta, A.C.M.F.; Mathews, P.D.; Madrid, R.R.M.; Rigoni, V.L.S.; Silva, E.R.; Mertins, O. Polyionic complexes of chitosan-N- arginine with alginate as pH responsive and mucoadhesive particles for oral drug delivery applications. Int. J. Biol. Macromol. 2020, 148, 550–564. [Google Scholar] [CrossRef] [PubMed]
- Mathews, P.D.; Patta, A.C.M.F.; Madrid, R.R.M.; Ramirez, C.A.B.; Pimenta, B.V.; Mertins, O. Efficient treatment of fish intestinal parasites applying membrane-penetrating oral drug delivery nanoparticle. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Madrid, R.R.M.; Mathews, P.D.; Patta, A.C.M.F.; Gonzales-Flores, A.P.; Ramirez, C.A.B.; Rigoni, V.L.S.; Tavares-Dias, M.; Mertins, O. Safety of oral administration of high doses of ivermectin by means of biocompatible polyelectrolytes formulation. Heliyon 2021, 7, e05820. [Google Scholar] [CrossRef]
- Madrid, R.R.M.; Mertins, O.; Tavares-Dias, M.; Flores-Gonzales, A.P.; Patta, A.C.M.F.; Ramirez, C.A.B.; Rigoni, V.L.S.; Mathews, P.D. High compliance and effective treatment of fish endoparasitic infections with oral drug delivery nanobioparticles: Safety of intestinal tissue and blood parameters. J. Fish Dis. 2021, 44, 1819–1829. [Google Scholar] [CrossRef]
- Silva, J.M.D. Estudos de síntese, conformação e atividade biológica de análogos do peptídeo antimicrobiano longipina. Master’s Thesis, Federal University of Sao Paulo, Sao Paulo, Brazil, 2017; 128p. [Google Scholar]
- Wittemann, A.; Ballauff, M. Interaction of proteins with linear polyelectrolytes and spherical polyelectrolyte brushes in aqueous solution. Phys. Chem. Chem. Phys. 2006, 8, 5269–5275. [Google Scholar] [CrossRef]
- Xu, X.; Angioletti-Uberti, S.; Lu, Y.; Dzubiella, J.; Ballauff, M. Interaction of proteins with polyelectrolytes: Comparison of theory to experiment. Langmuir 2019, 35, 5373–5391. [Google Scholar] [CrossRef]
- Kitagawa, H.; Ohge, H.; Yu, L.; Kayama, S.; Hara, T.; Kashiyama, S.; Kajihara, T.; Hisatsune, J.; Sueda, T.; Sugai, M. Aeromonas dhakensis is not a rare cause of Aeromonas bacteremia in Hiroshima, Japan. J. Infect. Chemother. 2020, 26, 316–320. [Google Scholar] [CrossRef]
- Chen, P.L.; Lamy, B.; Ko, W.C. Aeromonas dhakensis, an increasingly recognized human pathogen. Front. Microbiol. 2016, 7, 793. [Google Scholar] [CrossRef] [Green Version]
- Esteve, C.; Alcaide, E.; Blasco, M.D. Aeromonas hydrophila subsp. dhakensis isolated from feces, water and fish in Mediterranean Spain. Microbes Environ. 2012, 27, 367–373. [Google Scholar] [PubMed]
- Angelova, M.I.; Dimitrov, D.S. Liposome electroformation. Faraday Discus. Chem. Soc. 1986, 81, 303–311, 345–349. [Google Scholar] [CrossRef]
- Karatekin, E.; Sandre, O.; Guitouni, H.; Borghi, N.; Puech, P.H.; Brochard-Wyart, F. Cascades of transient pores in giant vesicles: Line tension and transport. Biophys. J. 2003, 84, 1734–1749. [Google Scholar] [CrossRef] [Green Version]
- Leomil, F.S.C.; Zoccoler, M.; Dimova, R.; Rike, K.A. PoET: Automated approach for measuring pore edge tension in giant unilamellar vesicles. Bioinform. Adv. 2021, 1, vbab037. [Google Scholar] [CrossRef]
- Mathews, P.D.; Patta, A.C.M.F.; Gonçalves, J.V.; Gama, G.S.; Garcia, I.T.S.; Mertins, O. Targeted drug delivery and treatment of endoparasites with biocompatible particles of pH-responsive structure. Biomacromolecules 2018, 19, 499–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carriero, M.M.; Maia, A.A.M.; Sousa, R.L.M.; Henrique-Silva, F. Characterization of a new strain of Aeromonas dhakensis isolated from diseased pacu fish (Piaractus mesopotamicus) in Brazil. J. Fish Dis. 2016, 39, 1285–1295. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Schatz, C.; Viton, C.; Delair, T.; Pichot, C.; Domard, A. Typical physicochemical behaviors of chitosan in aqueous solution. Biomacromolecules 2003, 4, 641–648. [Google Scholar] [CrossRef]
- Sonaje, K.; Chuang, E.-Y.; Lin, K.-J.; Yen, T.-C.; Su, F.-Y.; Tseng, M.T.; Sung, H.-W. Opening of epithelial tight junctions and enhancement of paracellular permeation by chitosan: Microscopic, ultrastructural, and computed-tomographic observations. Mol. Pharm. 2012, 9, 1271–1279. [Google Scholar] [CrossRef]
- Wang, J.; Kong, M.; Zhou, Z.; Yan, D.; Yu, X.; Cheng, X.; Feng, C.; Liu, Y.; Chen, X. Mechanism of surface charge triggered intestinal epithelial tight junction opening upon chitosan nanoparticles for insulin oral delivery. Carbohydr. Polym. 2017, 10, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Kamei, N.; Tamiwa, H.; Miyata, M.; Haruna, Y.; Matsumura, K.; Ogino, H.; Hirano, S.; Higashiyama, K.; Takeda-Morishita, M. Hydrophobic amino acid tryptophan shows promise as a potential absorption enhancer for oral delivery of biopharmaceuticals. Pharmaceutics 2018, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Artursson, P.; Lindmark, T.; Davis, S.S.; Illum, L. Pharmaceutical aspects of polymeric nanoparticles for oral drug delivery. J. Biomed. Nanotech. 2005, 1, 235–258. [Google Scholar]
- Artursson, P.; Lindmark, T.; Davis, S.S.; Illum, L. Effect of chitosan on the permeability of monolayers of intestinal epithelial cells (Caco-2). Pharm. Res. 1994, 11, 1358–1361. [Google Scholar] [CrossRef]
- Portet, T.; Dimova, R. A New method for measuring edge tensions and stability of lipid bilayers: Effect of membrane composition. Biophys. J. 2010, 99, 3264–3273. [Google Scholar] [CrossRef]
- Mertins, O.; Dimova, R. Insights on the interactions of chitosan with phospholipid vesicles. Part II: Membrane stiffening and pore formation. Langmuir 2013, 29, 14552–14559. [Google Scholar] [CrossRef] [PubMed]
- Tomas, J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012, 2012, 256261. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.-L.; Wu, C.-J.; Tsai, P.-J.; Tang, H.-J.; Chuang, Y.-C.; Lee, N.-Y.; Lee, C.-C.; Li, C.-W.; Li, M.-C.; Chen, C.-C.; et al. Virulence diversity among bacteremic Aeromonas isolates: Ex vivo, animal, and clinical evidences. PLoS ONE 2014, 9, e111213. [Google Scholar] [CrossRef]
- Mosser, T.; Talagrand-Reboul, E.; Colston, S.M.; Graf, J.; Figueras, M.J.; Jumas-Bilak, E.; Lamy, B. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 2015, 6, 1218. [Google Scholar] [CrossRef] [Green Version]
- Morinaga, Y.; Yanagihara, K.; Araki, N.; Harada, Y.; Yamada, K.; Akamatsu, N.; Matsuda, J.; Nishino, T.; Hasegawa, H.; Izumikawa, K.; et al. Clinical characteristics of seven patients with Aeromonas septicemia in a Japanese hospital. Tohoku J. Exp. Med. 2011, 225, 81–84. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-J.; Hsuan-Chen, W.; Hsueh, P.-R.; Chang, M.-C.; Tsai, P.-J.; Shih, H.-I.; Wang, H.-C.; Chou, P.-H.; Ko, W.-C. Clinical implications of species identification in monomicrobial Aeromonas bacteremia. PLoS ONE 2015, 10, e0117821. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Tsai, P.-J.; Chen, P.-L.; Wu, I.-C.; Lin, Y.-T.; Chen, Y.-H.; Wang, L.-R.; Ko, W.-C. Aeromonas aquariorum septicemia and enterocolitis in a cirrhotic patient. Diagn. Microbiol. Infect. Dis. 2012, 74, 406–408. [Google Scholar] [CrossRef] [PubMed]
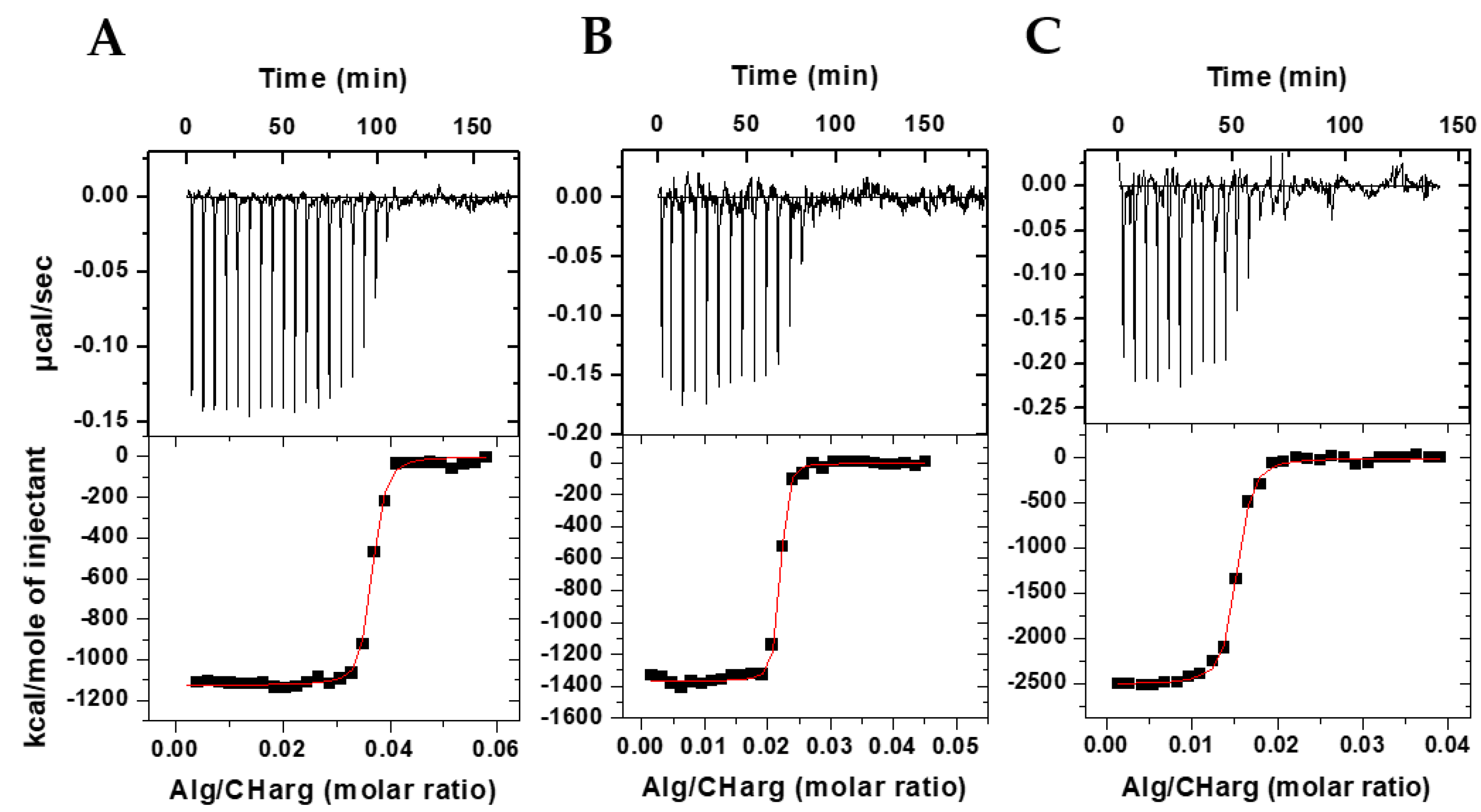

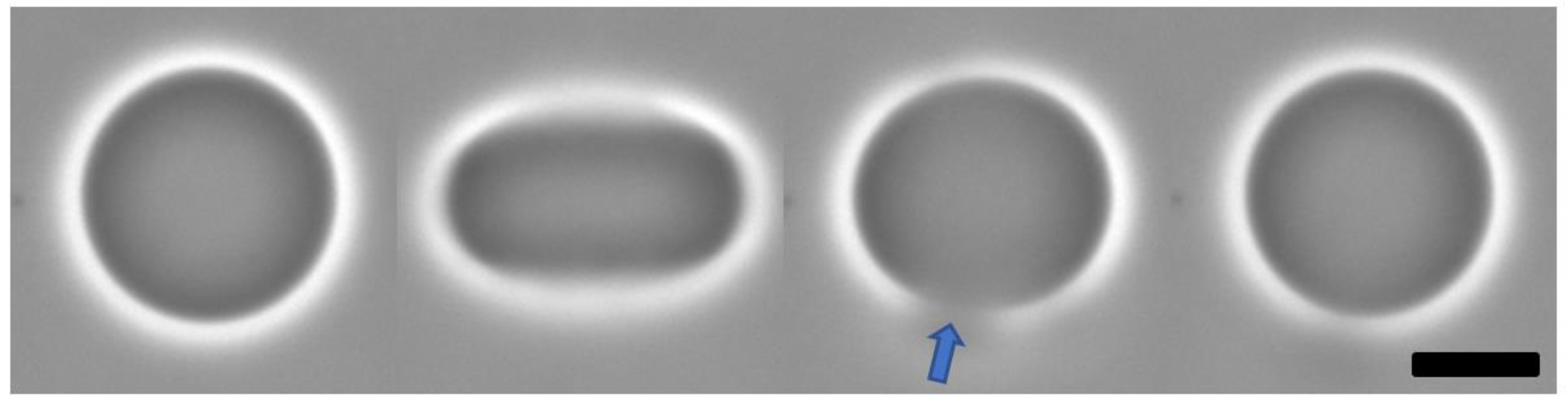
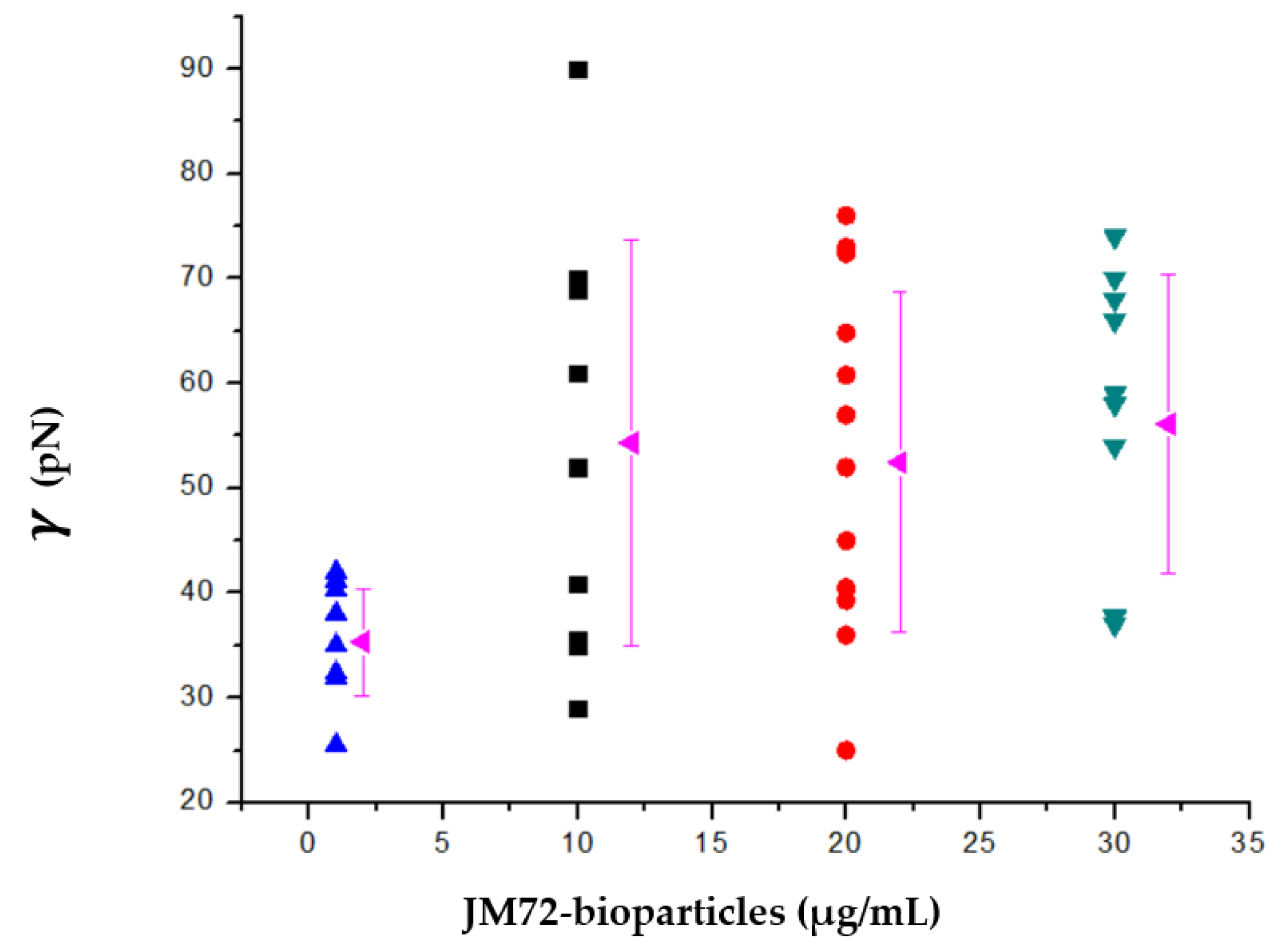

| Bioparticle | N | K (1010 M−1) | ΔH (kcal/mol) | ΔG (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|
| CHargAlg | 0.6290 ± 0.0050 | 0.029 ± 0.004 | −725 ± 8.9 | −13.93 | −711 |
| CHargAlg JM72 (2.5 µM) | 0.0368 ± 0.0002 | 2.95 ± 0.42 | −1125 ± 67 | −16.65 | −1108 |
| CHargAlg JM72 (7.5 µM) | 0.0225 ± 0.0007 | 5.53 ± 0.29 | −1369 ± 134 | −17.02 | −1352 |
| CHargAlg JM72 (15 µM) | 0.0147 ± 0.0003 | 4.17 ± 0.56 | −2514 ± 86 | −16.86 | −2497 |
| Buffer (pH) | Zeta Potential (mV) | Hydrodynamic Size (nm) | PDI |
|---|---|---|---|
| Citrate 2.5 | 17.6 ± 8.8 | 361 ± 60 (100%) | 0.52 |
| Acetate 4.5 | 19.1 ± 6.4 | 4139 ± 515 (57%) | 0.68 |
| 274 ± 26 (43%) | |||
| Phosphate 6.5 | 10.2 ± 2.9 | 867 ± 44 (83%) | 0.53 |
| 118 ± 18 (17%) | |||
| Phosphate 7.8 | 8.6 ± 6.4 | 568 ± 83 (61%) | 0.48 |
| 3421 ± 416 (39%) |
| Buffer (pH) | Zeta Potential (mV) | Hydrodynamic Size (nm) | PDI |
|---|---|---|---|
| Citrate 2.5 | 9.3 ± 6.7 | 101 ± 18 (100%) | 0.21 |
| Acetate 4.5 | 29.1 ± 9.6 | 538 ± 228 (62%) | 0.73 |
| 5463 ± 523 (30%) | |||
| 94 ± 15 (8%) | |||
| Phosphate 6.5 | −6.5 ± 5.6 | 383 ± 75 (74%) | 0.42 |
| 4328 ± 760 (26%) | |||
| Phosphate 7.8 | −9.8 ± 6.3 | 468 ± 91 (92%) | 0.43 |
| 2766 ± 728 (8%) |
| Buffer (pH) | Zeta Potential (mV) | Hydrodynamic Size (nm) | PDI |
|---|---|---|---|
| Citrate 2.5 | 31.1 ± 11.9 | 856 ± 110 (66%) | 0.47 |
| 157 ± 31 (23%) | |||
| 4999 ± 916 (11%) | |||
| Acetate 4.5 | 8.7 ± 4.5 | 278 ± 88 (65%) | 0.74 |
| 4624 ± 1010 (35%) | |||
| Phosphate 6.5 | 4.1 ± 7.3 | 912 ± 206 (89%) | 0.51 |
| 1147 ± 305 (11%) | |||
| Phosphate 7.8 | −5.9 ± 4.9 | 203 ± 90 (100%) | 0.69 |
| Buffer (pH) | Zeta Potential (mV) | Hydrodynamic Size (nm) | PDI |
|---|---|---|---|
| Citrate 2.5 | 2.4 ± 7.3 | 88 ± 31 (100%) | 0.47 |
| Acetate 4.5 | 17.4 ± 5.5 | 4799 ± 728 (65%) | 0.86 |
| 142 ± 65 (35%) | |||
| Phosphate 6.5 | 1.9 ± 5.4 | 334 ± 66 (100%) | 0.52 |
| Phosphate 7.8 | −2.6 ± 6.1 | 482 ± 110 (93%) | 0.51 |
| 7492 ± 1100 (7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramirez, C.A.B.; Carriero, M.M.; Leomil, F.S.C.; Moro de Sousa, R.L.; de Miranda, A.; Mertins, O.; Mathews, P.D. Complexation of a Polypeptide-Polyelectrolytes Bioparticle as a Biomaterial of Antibacterial Activity. Pharmaceutics 2022, 14, 2746. https://doi.org/10.3390/pharmaceutics14122746
Ramirez CAB, Carriero MM, Leomil FSC, Moro de Sousa RL, de Miranda A, Mertins O, Mathews PD. Complexation of a Polypeptide-Polyelectrolytes Bioparticle as a Biomaterial of Antibacterial Activity. Pharmaceutics. 2022; 14(12):2746. https://doi.org/10.3390/pharmaceutics14122746
Chicago/Turabian StyleRamirez, Carlos A. B., Mateus M. Carriero, Fernanda S. C. Leomil, Ricardo L. Moro de Sousa, Antonio de Miranda, Omar Mertins, and Patrick D. Mathews. 2022. "Complexation of a Polypeptide-Polyelectrolytes Bioparticle as a Biomaterial of Antibacterial Activity" Pharmaceutics 14, no. 12: 2746. https://doi.org/10.3390/pharmaceutics14122746





