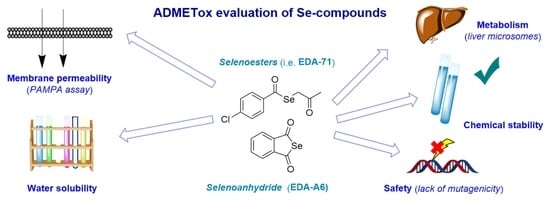
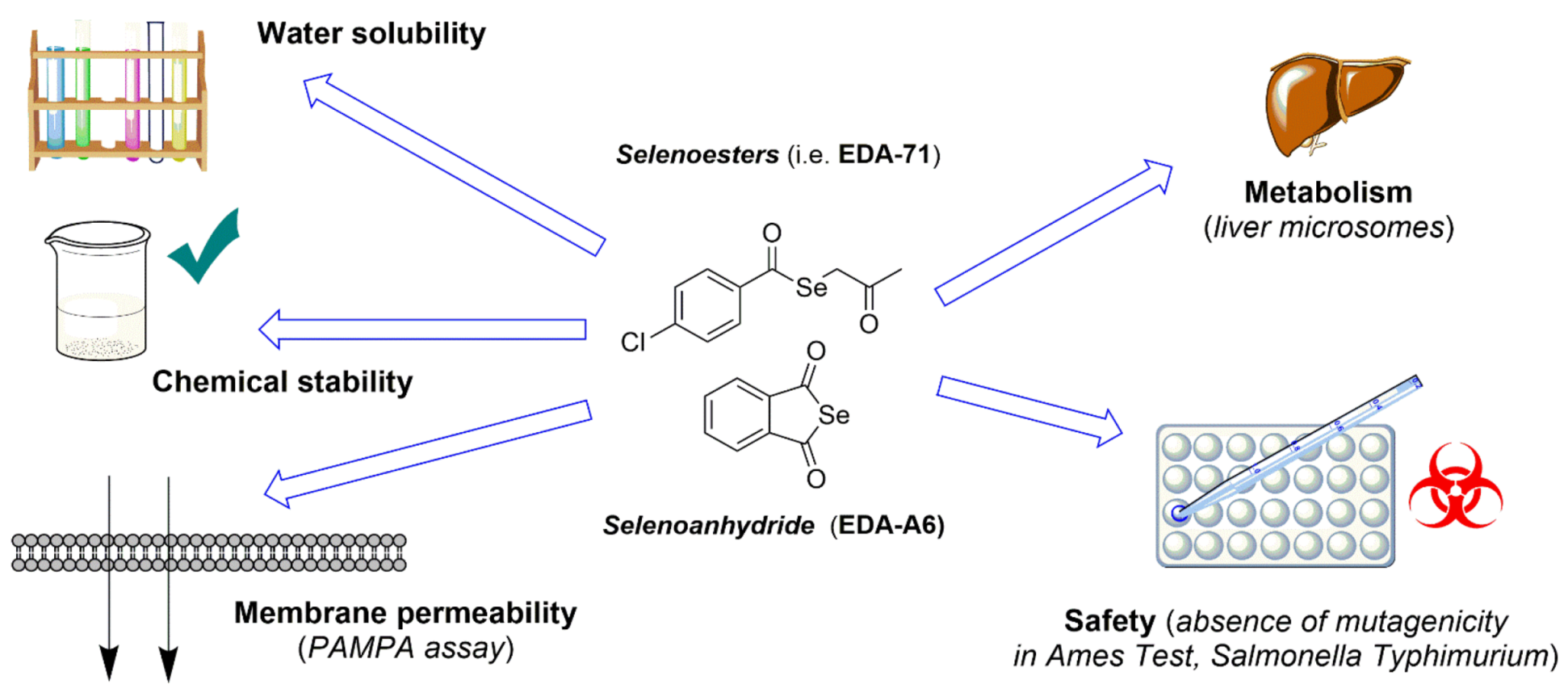
Pharmaceutical and Safety Profile Evaluation of Novel Selenocompounds with Noteworthy Anticancer Activity
Abstract
:1. Introduction
2. Materials and Methods
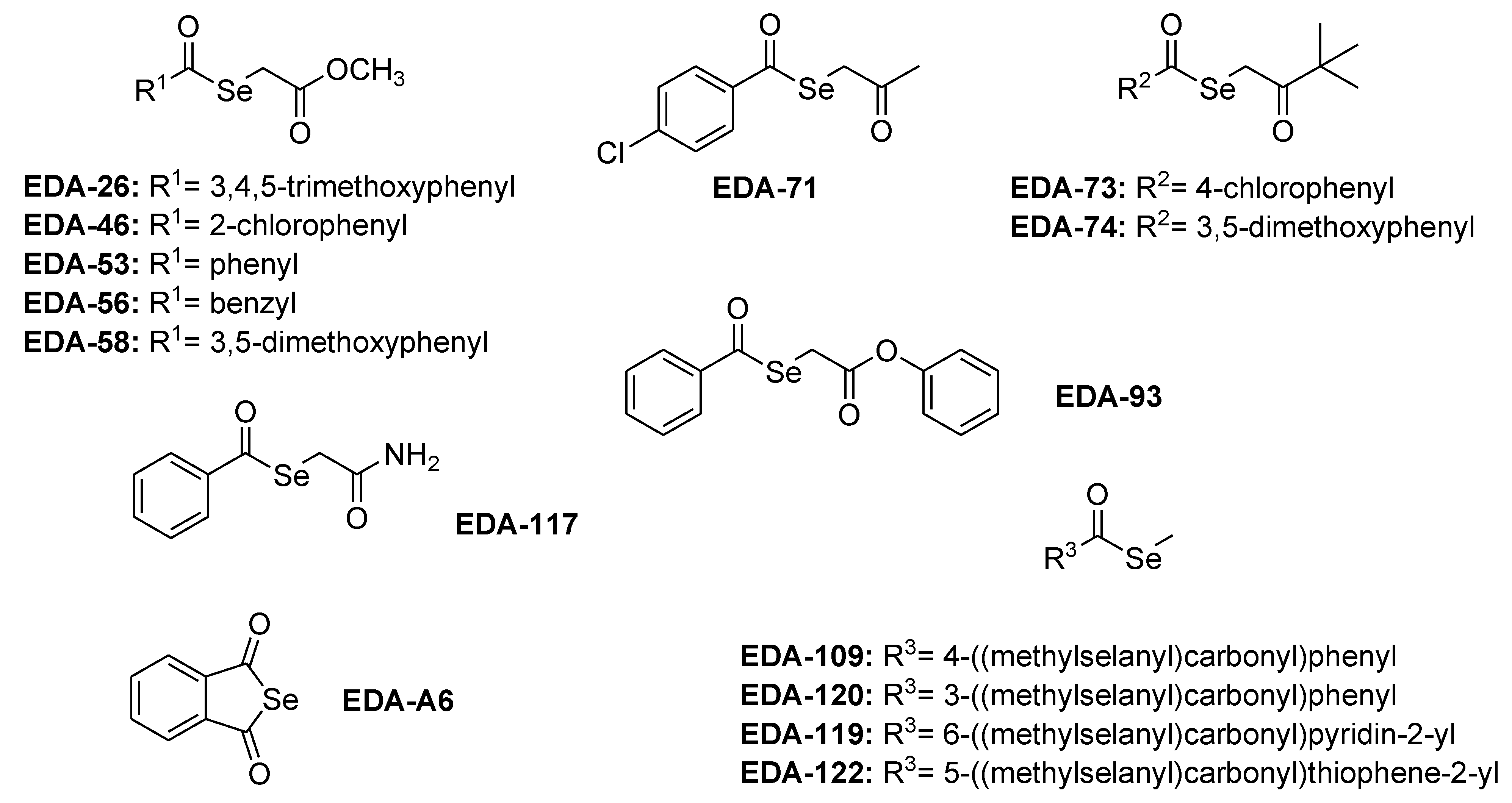
2.1. Tested Organic Selenocompounds
2.2. Reagents and Equipment
2.3. Determination of the Aqueous Solubility
2.4. Chemical Stability in the Acidic and Alkaline Environments
2.5. PAMPA Test: Membrane Permeability Evaluation
2.6. Metabolic Stability Evaluation
2.6.1. In Silico Metabolic Stability Simulation (MetaSite)
2.6.2. In Vitro Microsomal Biotransformation Tests
2.7. Safety Profile Evaluation—Ames Microplate Mutagenicity Assay
3. Results and Discussion
3.1. Determination of Aqueous Solubility
3.2. Chemical Stability in the Acidic and Alkaline Environments
3.3. PAMPA Test: Membrane Permeability Evaluation
3.4. Metabolic Stability Evaluation
3.4.1. In Silico Metabolic Stability Simulation (MetaSite)
3.4.2. In Vitro Microsomal Biotransformation Tests
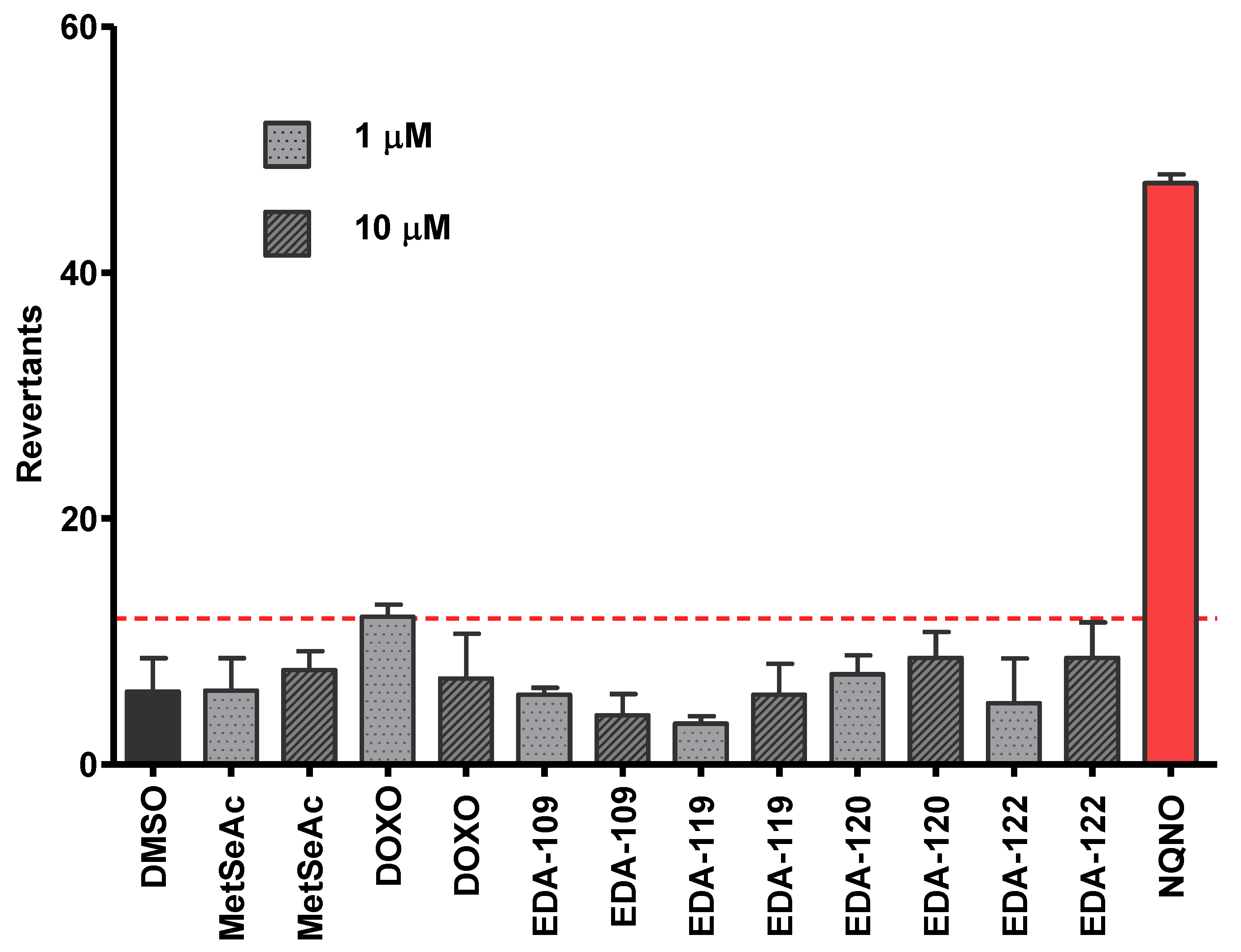
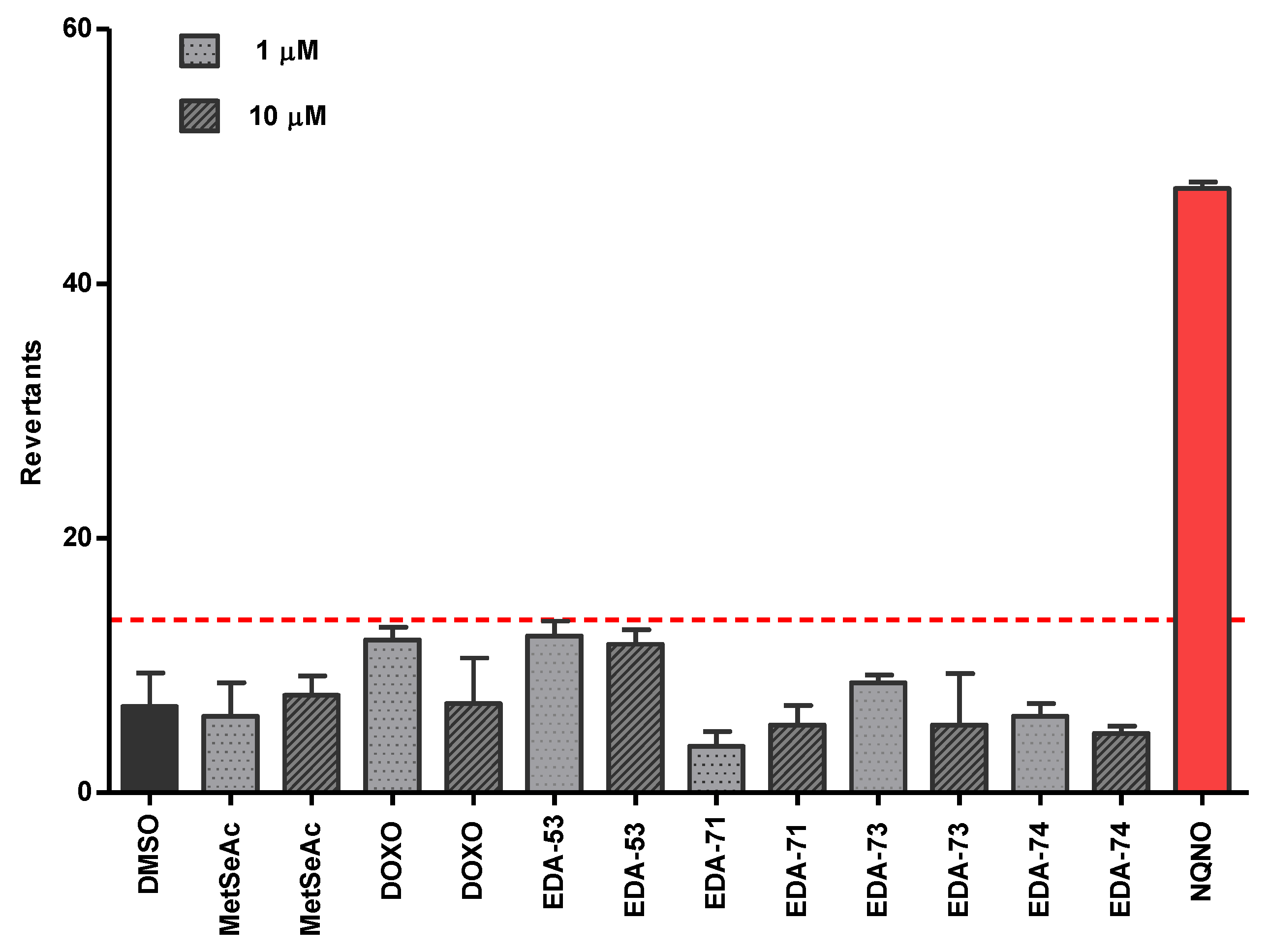
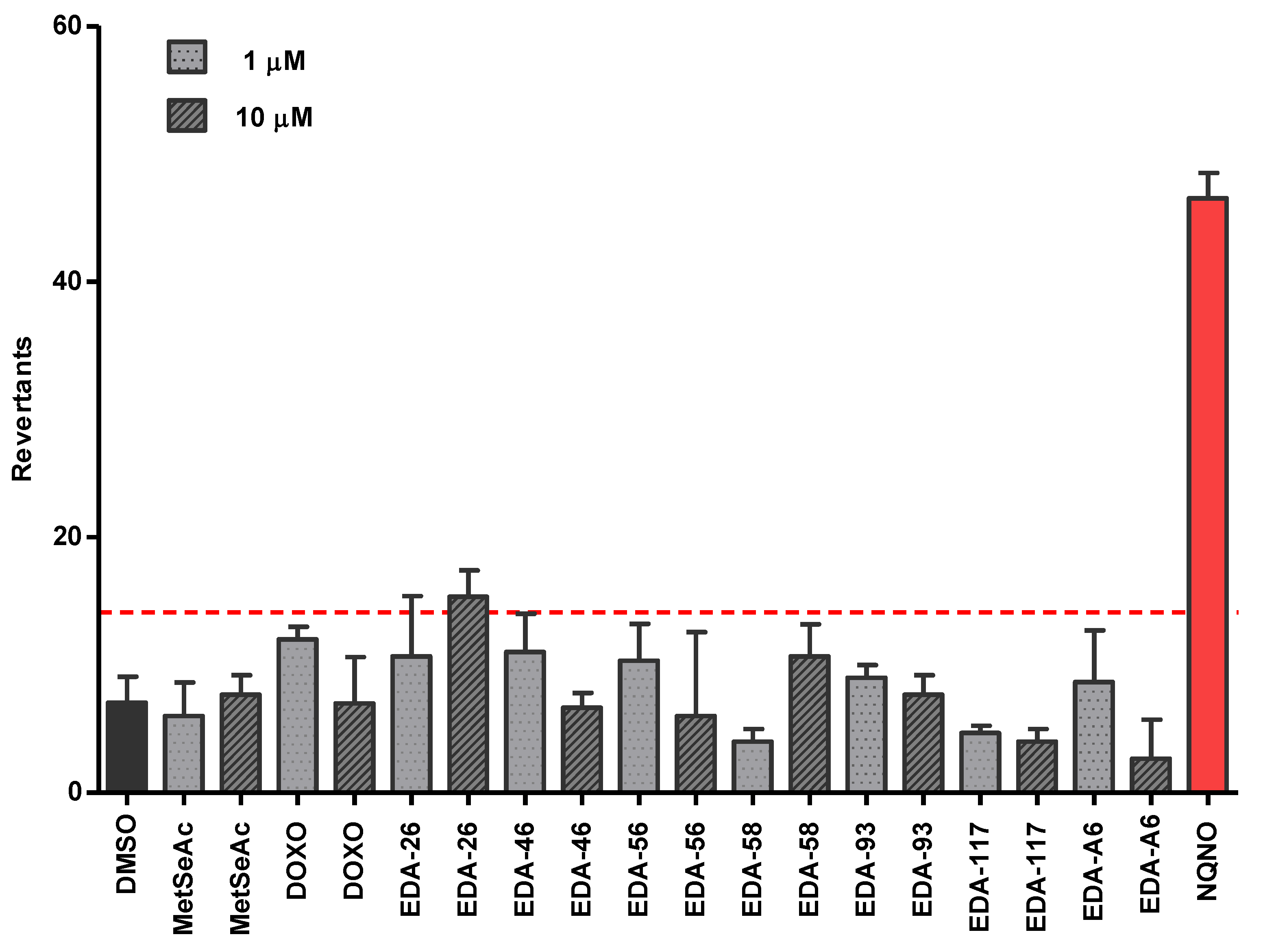
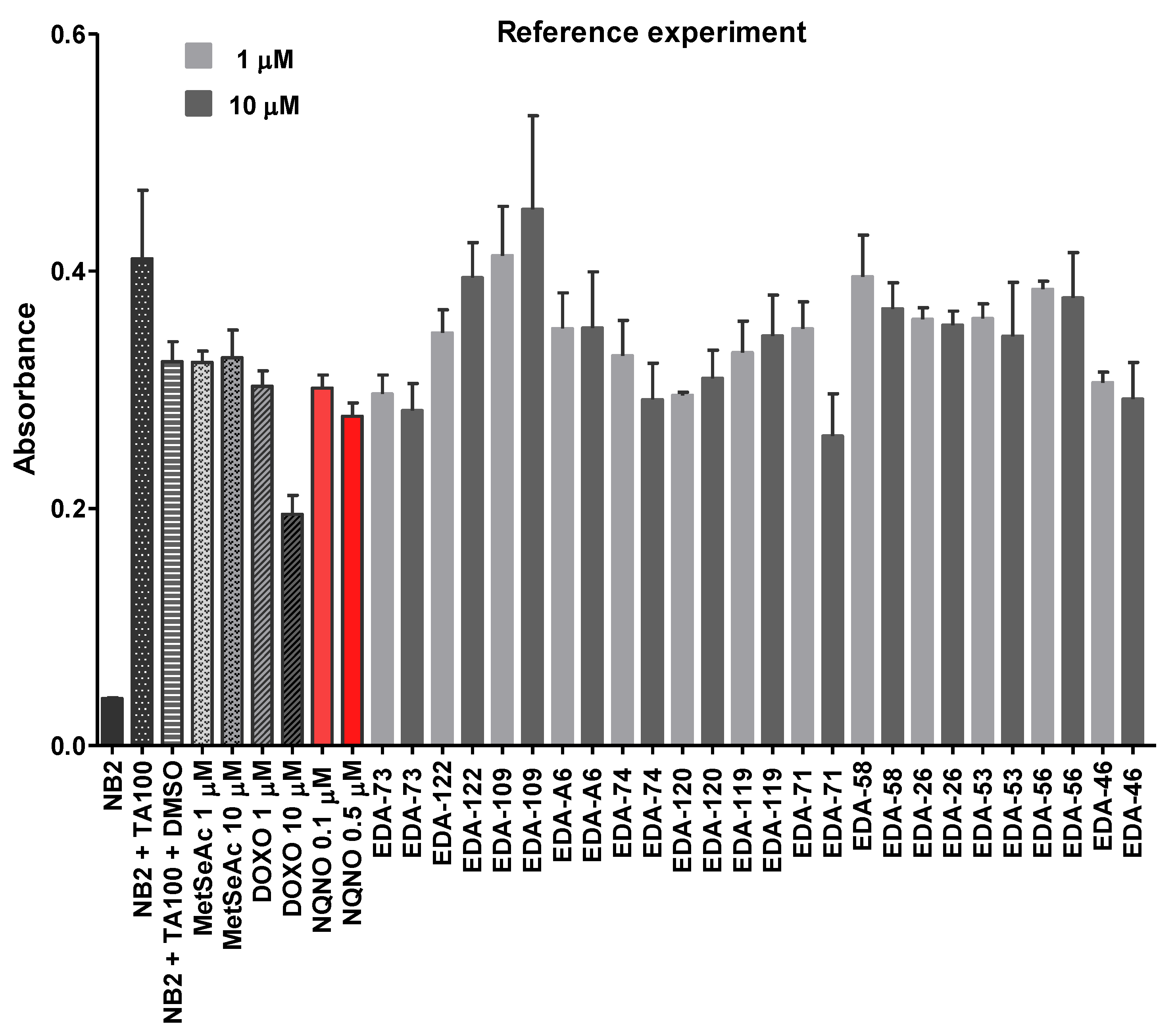
3.5. Evaluation of the Mutagenicity through the Ames Test
3.5.1. Safety Profile In Silico Results
3.5.2. Safety Profile In Vitro Results in Ames Microplate Mutagenicity Assay
3.5.3. Reference Experiment Results—The Impact of Tested Selenocompounds on the Salmonella Typhimurium Growth
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rayman, M.P. Selenium intake, status, and health: A complex relationship. Hormones 2020, 19, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogueira, C.W.; Rocha, J.B. Toxicology and pharmacology of selenium: Emphasis on synthetic organoselenium compounds. Arch. Toxicol. 2011, 85, 1313–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Y.; Zhang, H.; Liu, J. Safety Assessment and Comparison of Sodium Selenite and Bioselenium Obtained from Yeast in Mice. Biomed. Res. Int. 2017, 2017, 3980972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radomska, D.; Czarnomysy, R.; Radomski, D.; Bielawska, A.; Bielawski, K. Selenium as a Bioactive Micronutrient in the Human Diet and Its Cancer Chemopreventive Activity. Nutrients 2021, 13, 1649. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Peyroche, G.; Saveanu, C.; Dauplais, M.; Lazard, M.; Beuneu, F.; Decourty, L.; Malabat, C.; Jacquier, A.; Blanquet, S.; Plateau, P. Sodium selenide toxicity is mediated by O2-dependent DNA breaks. PLoS ONE 2012, 7, e36343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Nitteranon, V.; Guo, S.; Qiu, P.; Wu, X.; Li, F.; Xiao, H.; Hu, Q.; Parkin, K.L. Organoselenium compounds modulate extracellular redox by induction of extracellular cysteine and cell surface thioredoxin reductase. Chem. Res. Toxicol. 2013, 26, 456–464. [Google Scholar] [CrossRef]
- Brozmanová, J.; Mániková, D.; Vlčková, V.; Chovanec, M. Selenium: A double-edged sword for defense and offence in cancer. Arch. Toxicol. 2010, 84, 919–938. [Google Scholar] [CrossRef]
- Lee, K.H.; Jeong, D. Bimodal actions of selenium essential for antioxidant and toxic pro-oxidant activities: The selenium paradox (Review). Mol. Med. Rep. 2012, 5, 299–304. [Google Scholar]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef]
- Jaworska, K.; Gupta, S.; Durda, K.; Muszyńska, M.; Sukiennicki, G.; Jaworowska, E.; Grodzki, T.; Sulikowski, M.; Waloszczyk, P.; Wójcik, J.; et al. A low selenium level is associated with lung and laryngeal cancers. PLoS ONE 2013, 8, e59051. [Google Scholar] [CrossRef]
- Bartolini, D.; Sancineto, L.; Fabro de Bem, A.; Tew, K.D.; Santi, C.; Radi, R.; Toquato, P.; Galli, F. Selenocompounds in Cancer Therapy: An Overview. Adv. Cancer Res. 2017, 136, 259–302. [Google Scholar]
- Radomska, D.; Czarnomysy, R.; Radomski, D.; Bielawski, K. Selenium Compounds as Novel Potential Anticancer Agents. Int. J. Mol. Sci. 2021, 22, 1009. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Álvarez-Pérez, M.; Marć, M.A.; Salardón-Jiménez, N.; Handzlik, J.; Domínguez-Álvarez, E. The Anticancer and Chemopreventive Activity of Selenocyanate-Containing Compounds. Curr. Pharmacol. Rep. 2018, 4, 468–481. [Google Scholar] [CrossRef]
- Chuai, H.; Zhang, S.Q.; Bai, H.; Li, J.; Wang, Y.; Sun, J.; Wen, E.; Zhang, J.; Xin, M.K. Small molecule selenium-containing compounds: Recent development and therapeutic applications. Eur. J. Med. Chem. 2021, 223, 113621. [Google Scholar] [CrossRef]
- Fernandes, A.P.; Gandin, V. Selenium compounds as therapeutic agents in cancer. Biochim. Biophys. Acta 2015, 1850, 1642–1660. [Google Scholar] [CrossRef]
- Kim, S.J.; Choi, M.C.; Park, J.M.; Chung, A.S. Antitumor Effects of Selenium. Int. J. Mol. Sci. 2021, 22, 11844. [Google Scholar] [CrossRef]
- Domínguez-Álvarez, E.; Plano, D.; Font, M.; Calvo, A.; Prior, C.; Jacob, C.; Palop, J.A.; Sanmartín, C. Synthesis and antiproliferative activity of novel selenoester derivatives. Eur. J. Med. Chem. 2014, 73, 153–166. [Google Scholar] [CrossRef]
- Gajdács, M.; Spengler, G.; Sanmartín, C.; Marć, M.A.; Handzlik, J.; Domínguez-Álvarez, E. Selenoesters and selenoanhydrides as novel multidrug resistance reversing agents: A confirmation study in a colon cancer MDR cell line. Bioorg. Med. Chem. Lett. 2017, 27, 797–802. [Google Scholar] [CrossRef] [Green Version]
- Kharma, A.; Misak, A.; Marian Grman, G.; Brezova, V.; Kurakova, L.; Baráth, P.; Jacob, C.; Chovanec, M.; Ondrias, K.; Domínguez-Álvarez, E. Release of reactive selenium species from phthalic selenoanhydride in the presence of hydrogen sulfide and glutathione with implications for cancer research. New J. Chem. 2019, 43, 11771–11783. [Google Scholar] [CrossRef] [Green Version]
- Spengler, G.; Kincses, A.; Mosolygó, T.; Marć, M.A.; Nové, M.; Gajdács, M.; Sanmartín, C.; McNeil, H.E.; Blair, J.M.A.; Domínguez-Álvarez, E. Antiviral, Antimicrobial and Antibiofilm Activity of Selenoesters and Selenoanhydrides. Molecules 2019, 24, 4264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosolygó, T.; Kincses, A.; Csonka, A.; Tönki, Á.S.; Witek, K.; Sanmartín, C.; Marć, M.A.; Handzlik, J.; Kieć-Kononowicz, K.; Domínguez-Álvarez, E.; et al. Selenocompounds as Novel Antibacterial Agents and Bacterial Efflux Pump Inhibitors. Molecules 2019, 24, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, L.; Kerns, E.H.; Carter, G. Drug-like property concepts in pharmaceutical design. Curr. Pharm. Des. 2009, 15, 2184–2194. [Google Scholar] [CrossRef] [PubMed]
- Kerns, E.H.; Di, L. Drug-like Properties: Concepts, Structure, Design and Methods; Academic Press: Cambridge, MA, USA, 2008; ISBN 978-0-1236-9520-8. [Google Scholar]
- Kong, W.M.; Chik, Z.; Mohamed, Z.; Alshawsh, M.A. Physicochemical Characterization of Mitragyna speciosa Alkaloid Extract and Mitragynine Using In Vitro High Throughput Assays. Comb. Chem. High. Throughput Screen 2017, 20, 796–803. [Google Scholar] [CrossRef]
- Alam, S.; Khan, F. 3D-QSAR, Docking, ADME/Tox studies on Flavone analogs reveal anticancer activity through Tankyrase inhibition. Sci. Rep. 2019, 9, 5414. [Google Scholar] [CrossRef] [PubMed]
- DeMarini, D.M.; Shelton, M.L.; Warren, S.H.; Ross, T.M.; Shim, J.Y.; Richard, A.M.; Pegram, R.A. Glutathione S-transferase-mediated induction of GC-->AT transitions by halomethanes in Salmonella. Environ. Mol. Mutagen. 1997, 30, 440–447. [Google Scholar] [CrossRef]
- DeMarini, D.M.; Abu-Shakra, A.; Felton, C.F.; Patterson, K.S.; Shelton, M.L. Mutation spectra in salmonella of chlorinated, chloraminated, or ozonated drinking water extracts: Comparison to MX. Environ. Mol. Mutagen. 1995, 26, 270–285. [Google Scholar] [CrossRef]
- Marć, M.A.; Domínguez-Álvarez, E.; Słoczyńska, K.; Żmudzki, P.; Chłoń-Rzepa, G.; Pękala, E. In Vitro Biotransformation, Safety, and Chemopreventive Action of Novel 8-Methoxy-Purine-2,6-Dione Derivatives. Appl. Biochem. Biotechnol. 2018, 184, 124–139. [Google Scholar] [CrossRef] [Green Version]
- Mortelmans, K.; Zeiger, E. The Ames Salmonella/microsome mutagenicity assay. Mutat. Res. 2000, 455, 29–60. [Google Scholar] [CrossRef]
- Zeiger, E. Bacterial mutation assays. Methods Mol. Biol. 2013, 1044, 3–26. [Google Scholar]
- Ames, B.N.; Lee, F.D.; Durston, W.E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc. Natl. Acad. Sci. USA 1973, 70, 782–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Southan, C.; Stracz, A. Extracting and connecting chemical structures from text sources using chemicalize.org. J. Cheminform. 2013, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; Reich, J.R.; Rufener, C. OSIRIS, an entirely in-house developed drug discovery informatics system. J. Chem. Inf. Model 2009, 49, 232–246. [Google Scholar] [CrossRef]
- Vollmann, K.; Qurishi, R.; Hockemeyer, J.; Müller, C.E. Synthesis and properties of a new water-soluble prodrug of the adenosine A 2A receptor antagonist MSX-2. Molecules 2008, 13, 348–359. [Google Scholar] [CrossRef] [Green Version]
- Żesławska, E.; Kucwaj-Brysz, K.; Kincses, A.; Spengler, G.; Szymańska, E.; Czopek, A.; Marć, M.A.; Kaczor, A.; Nitek, W.; Domínguez-Álvarez, E.; et al. An insight into the structure of 5-spiro aromatic derivatives of imidazolidine-2,4-dione, a new group of very potent inhibitors of tumor multidrug resistance in T-lymphoma cells. Bioorg. Chem. 2021, 109, 104735. [Google Scholar] [CrossRef]
- Nasim, M.J.; Witek, K.; Kincses, A.; Abdin, A.Y.; Żesławska, E.; Marć, M.A.; Gajdács, M.; Spengler, G.; Nitek, W.; Latacz, G.; et al. Pronounced activity of aromatic selenocyanates against multidrug resistant ESKAPE bacteria. New J. Chem. 2019, 43, 6021–6031. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Murawski, A.; Patel, K.; Crespi, C.L.; Balimane, P.V. A novel design of artificial membrane for improving the PAMPA model. Pharm. Res. 2008, 25, 1511–1520. [Google Scholar] [CrossRef]
- Bujard, A.; Voirol, H.; Carrupt, P.A.; Schappler, J. Modification of a PAMPA model to predict passive gastrointestinal absorption and plasma protein binding. Eur. J. Pharm. Sci. 2015, 77, 273–278. [Google Scholar]
- Latacz, G.; Lubelska, A.; Jastrzębska-Więsek, M.; Partyka, A.; Sobiło, A.; Olejarz, A.; Kucwaj-Brysz, K.; Satała, G.; Bojarski, A.J.; Wesołowska, A.; et al. In the search for a lead structure among series of potent and selective hydantoin 5-HT7 R agents: The drug-likeness in vitro study. Chem. Biol. Drug Res. 2017, 90, 1295–1306. [Google Scholar] [CrossRef]
- Łażewska, D.; Więcek, M.; Ner, J.; Kamińska, K.; Kottke, T.; Schwed, J.S.; Zygmunt, M.; Karcz, T.; Olejarz, A.; Kuder, K.; et al. Aryl -1,2,5-triazine derivatives as histaminę H4 receptor ligands. Eur. J. Med. Chem. 2014, 83, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Kucwaj-Brysz, K.; Warszycki, D.; Podlewska, S.; Witek, J.; Witek, K.; González Izquierdo, A.; Satała, G.; Loza, M.I.; Lubelska, A.; Latacz, G.; et al. Rational design in search for 5-phenylhydantoin selective 5-HT7R antagonists. Molecular modeling, synthesis and biological evaluation. Eur. J. Med. Chem. 2016, 112, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Kamiński, K.; Zagaja, M.; Łuszczki, J.J.; Rapacz, A.; Andres-Mach, M.; Latacz, G.; Kieć-Kononowicz, K. Design, Synthesis, and Anticonvulsant Activity of New Hybrid Compounds Derived from 2-(2,5-Dioxopyrrolidin-1-yl)propanamides and 2-(2,5-Dioxopyrrolidin-1-yl)butanamides. J. Med. Chem. 2015, 58, 5274–5286. [Google Scholar] [CrossRef] [PubMed]
- Sadek, B.; Saad, A.; Latacz, G.; Kuder, K.; Olejarz, A.; Karcz, T.; Stark, H.; Kieć-Kononowicz, K. Non-imidazole-based histamine H3 receptor antagonists with anticonvulsant activity in different seizure models in male adult rats. Drug Des. Dev. Ther. 2016, 10, 3879–3898. [Google Scholar] [CrossRef] [Green Version]
- Witek, K.; Nasim, M.J.; Bischoff, M.; Gaupp, R.; Arsenyan, P.; Vasiljeva, J.; Marć, M.A.; Olejarz, A.; Latacz, G.; Kieć-Kononowicz, K.; et al. Selenazolinium Salts as “Small Molecule Catalysts” with High Potency against ESKAPE Bacterial Pathogens. Molecules 2017, 22, E2174. [Google Scholar] [CrossRef] [Green Version]
- Flückiger-Isler, S.; Baumeister, M.; Braun, K.; Gervais, V.; Hasler-Nguyen, H.; Reimann, R.; van Gompel, J.; Wunderlich, H.-G.; Engelhardt, G. Assessment of the performance of the Ames IITM assay: A collaborative study with 19 coded compounds. Mutat. Res. 2004, 558, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Kamber, M.; Fluckinger-Isler, S.; Engelhardt, G.; Jaeckh, R.; Zeiger, E. Comparison of the Ames II and traditional Ames test responses with respect to mutagenicity, strain specificities, need for metabolism and correlation with rodent carcinogenicity. Mutagenesis 2009, 24, 359–366. [Google Scholar] [CrossRef] [Green Version]
- de Umbuzeiro, G.A.; Rech, C.M.; Correia, S.; Bergamasco, A.M.; Cardenette, G.H.; Fluckiger-Isler, S.; Kamber, M. Comparison of the Salmonella/Microsome Microsuspension Assay with the new Microplate Fluctuation Protocol for Testing the Mutagenicity of Environmental Samples. Environ. Mol. Mutagen. 2010, 51, 31–38. [Google Scholar] [CrossRef]
- Fronza, G.; Campomenosi, P.; Iannone, R.; Abbondandolo, A. The 4-nitroquinoline 1-oxide mutational spectrum in single stranded DNA is characterized by guanine to pyrimidine transversions. Nucleic Acids Res. 1992, 20, 1283–1287. [Google Scholar] [CrossRef] [Green Version]
- U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER). Guidance for Industry: Waiver of In Vivo Bioavailability and Bioequivalence Studies for Immediate Release Solid Oral Dosage Forms Based on a Biopharmaceutics Classification System; U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER): Beltsville, MD, USA, 2015. [Google Scholar]
- CPMP/EWP/QWP/1401/98; Guideline on the Investigation of Bioequivalence; European Medicines Agency (EMA): Amsterdam, The Netherlands, 2008.
- Espuelas, S.; Plano, D.; Nguewa, P.; Font, M.; Palop, J.A.; Irache, J.M.; Sanmartín, C. Innovative lead compounds and formulation strategies as newer kinetoplastid therapies. Curr. Med. Chem. 2012, 19, 4259–4288. [Google Scholar] [CrossRef]
- Ruberte, A.C.; González-Gaitano, G.; Sharma, A.K.; Aydillo, C.; Encío, I.; Sanmartín, C.; Plano, D. New Formulation of a Methylseleno-Aspirin Analog with Anticancer Activity towards Colon Cancer. Int. J. Mol. Sci. 2020, 21, 9017. [Google Scholar] [CrossRef] [PubMed]
- Berntssen, M.H.G.; Sundal, T.K.; Olsvik, P.A.; Amlund, H.; Rasinger, J.D.; Sele, V.; Hamre, K.; Hillestad, M.; Buttle, L.; Ørnsrud, R. Sensitivity and toxic mode of action of dietary organic and inorganic selenium in Atlantic salmon (Salmo salar). Aquat. Toxicol. 2017, 192, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, M.; Inoue, H.; Kohno, M.; Saito, M.; Tsuge, S.; Shimizu, S.; Ishida, A.; Ishibashi, O.; Inui, T. Human Lipocalin-Type Prostaglandin D Synthase-Based Drug Delivery System for Poorly Water-Soluble Anti-Cancer Drug SN-38. PLoS ONE 2015, 10, e0142206. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, H.; Komasaka, T.; Tomari, T.; Kitano, Y.; Takekawa, K. Nanosuspension formulations of poorly water-soluble compounds for intravenous administration in exploratory toxicity studies: In vitro and in vivo evaluation. J. Appl. Toxicol. 2016, 36, 1259–1267. [Google Scholar] [CrossRef]
- Petit, C.; Bujard, A.; Skalicka-Woźniak, K.; Cretton, S.; Houriet, J.; Christen, P.; Carrupt, P.A.; Wolfender, J.L. Prediction of the Passive Intestinal Absorption of Medicinal Plant Extract Constituents with the Parallel Artificial Membrane Permeability Assay (PAMPA). Planta Med. 2016, 82, 424–431. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Okochi, H.; Benet, L.Z.; Zhai, S.D. Sotalol Permeability in Cultured-Cell, Rat Intestine, and PAMPA System. Pharm. Res. 2012, 29, 1768–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markovic, B.D.; Vladimirov, S.M.; Cudina, O.A.; Odovic, J.V.; Karljikovic-Rajic, K.D. A PAMPA Assay as Fast Predictive Model of Passive Human Skin Permeability of New Synthesized Corticosteroid C-21 Esters. Molecules 2012, 17, 480–491. [Google Scholar] [CrossRef] [Green Version]
- Hallifax, D.; Turlizzi, E.; Zanelli, U.; Houston, J.B. Clearance-dependent underprediction of in vivo intrinsic clearance from human hepatocytes: Comparison with permeabilities from artificial membrane (PAMPA) assay, in silico and caco-2 assay, for 65 drugs. Eur. J. Pharm. Sci. 2012, 45, 570–574. [Google Scholar] [CrossRef]
- Serra, H.; Mendes, T.; Bronze, M.R.; Simplício, A.L. Prediction of intestinal absorption and metabolism of pharmacologically active flavones and flavanones. Bioorg. Med. Chem. 2008, 16, 4009–4018. [Google Scholar] [CrossRef]
- Takahashi, K.; Suzuki, N.; Ogra, Y. Bioavailability Comparison of Nine Bioselenocompounds In Vitro and In Vivo. Int. J. Mol. Sci. 2017, 18, 506. [Google Scholar] [CrossRef] [Green Version]
- Mukkavilli, R.; Yang, C.; Singh Tanwar, R.; Ghareeb, A.; Luthra, L.; Aneja, R. Absorption, Metabolic Stability, and Pharmacokinetics of Ginger Phytochemicals. Molecules 2017, 22, 553. [Google Scholar] [CrossRef] [PubMed]
- Letelier, M.E.; Izquierdo, P.; Godoy, L.; Lepe, A.M.; Faúndez, M. Liver Microsomal Biotransformation of Nitro-aryl Drugs: Mechanism for Potential Oxidative Stress Induction. J. Appl. Toxicol. 2004, 24, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Fasinu, P.; Bouic, P.J.; Rosenkranz, B. Liver-based in vitro technologies for drug biotransformation studies—A review. Curr. Drug Metab. 2012, 13, 215–224. [Google Scholar] [CrossRef]
- Yilmazer, M.; Stevens, J.F.; Deinzer, M.L.; Buhler, D.R. In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes. Drug Metab. Dispos. 2001, 29, 223–231. [Google Scholar] [PubMed]
- Xiong, H.; Turner, K.C.; Ward, E.S.; Jansen, P.L.; Brouwer, K.L. Altered hepatobiliary disposition of acetaminophen glucuronide in isolated perfused livers from multidrug resistance-associated protein 2-deficient TR(-) rats. J. Pharmacol. Exp. Ther. 2000, 295, 512–518. [Google Scholar] [PubMed]
- Asha, S.; Vidyavathi, M. Cunninghamella-a microbial model for drug metabolism studies-a review. Biotechnol. Adv. 2009, 27, 16–29. [Google Scholar] [CrossRef]
- Zagrodzki, P.; Paśko, P.; Domínguez-Álvarez, E.; Salardón-Jiménez, N.; Sevilla-Hernández, C.; Sanmartín, C.; Bierła, K.; Łobiński, R.; Szpunar, J.; Handzlik, J.; et al. Synthesis of novel organic selenium compounds and speciation of their metabolites in biofortified kale sprouts. Microchem. J. 2022, 172, 106962. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, Y.; Zhong, J.; Pan, Y.; Cai, S.; Xu, J. Toxicological profile and safety pharmacology of a single dose of fibroblast activation protein-α-based doxorubicin prodrug: In-vitro and in-vivo evaluation. Anticancer. Drugs 2018, 29, 253–261. [Google Scholar] [CrossRef]
- MacLachlan, T.K.; Milton, M.N.; Turner, O.; Tukov, F.; Choi, V.W.; Penraat, J.; Delmotte, M.H.; Michaut, L.; Jaffee, B.D.; Bigelow, C.E. Nonclinical Safety Evaluation of scAAV8-RLBP1 for Treatment of RLBP1 Retinitis Pigmentosa. Mol. Ther. Methods Clin. Dev. 2017, 22, 105–120. [Google Scholar] [CrossRef] [Green Version]
- MacArthur Clark, J. The 3Rs in research: A contemporary approach to replacement, reduction and refinement. Br. J. Nutr. 2017, 120, S1–S7. [Google Scholar] [CrossRef]
- Ragazzo, P.; Feretti, D.; Monarca, S.; Dominici, L.; Ceretti, E.; Viola, G.; Piccolo, V.; Chiucchini, N.; Villarini, M. Evaluation of cytotoxicity, genotoxicity, and apoptosis of wastewater before and after disinfection with performic acid. Water Res. 2017, 116, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Shigenaga, M.K.; Gold, L.S. DNA lesions, inducible DNA repair, and cell division: Three key factors in mutagenesis and carcinogenesis. Environ. Health Perspect. 1993, 101, 35–44. [Google Scholar] [PubMed] [Green Version]
- Słoczyńska, K.; Powroźnik, B.; Pękala, E.; Waszkielewicz, A.M. Antimutagenic compounds and their possible mechanisms of action. J. Appl. Genet. 2014, 55, 273–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975, 31, 217–233. [Google Scholar] [CrossRef]
- Szemerédi, N.; Kincses, A.; Rehorova, K.; Hoang, L.; Salardón-Jiménez, N.; Sevilla-Hernández, C.; Viktorová, J.; Domínguez-Álvarez, E.; Spengler, G. Ketone- and Cyano-Selenoesters to Overcome Efflux Pump, Quorum-Sensing, and Biofilm-Mediated Resistance. Antibiotic 2020, 9, 896. [Google Scholar] [CrossRef]
- Kandaş, N.O.; Randolph, C.; Bosland, M.C. Differential effects of selenium on benign and malignant prostate epithelial cells: Stimulation of LNCaP cell growth by noncytotoxic, low selenite concentrations. Nutr. Cancer 2009, 61, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Atten, M.J.; Attar, B.M.; Holian, O. Selenomethionine stimulates MAPK (ERK) phosphorylation, protein oxidation, and DNA synthesis in gastric cancer cells. Nutr. Cancer 2004, 49, 184–190. [Google Scholar] [CrossRef]
- Benassi, J.C.; Barbosa, F.A.R.; Candiotto, G.; Grinevicius, V.M.A.S.; Filho, D.W.; Braga, A.L.; Pedrosa, R.C. Docking and molecular dynamics predicted B-DNA and dihydropyrimidinone selenoesters interactions elucidating antiproliferative effects on breast adenocarcinoma cells. J. Biomol. Struct. Dyn. 2021; inpress. [Google Scholar] [CrossRef]
- Gajdács, M.; Handzlik, J.; Sanmartín, C.; Domínguez-Álvarez, E.; Spengler, G. Rákellenes és efflux pumpa gátló hatású szelénvegyületek ADME tulajdonságainak becslése számítógépes módszerrel [Prediction of ADME properties for selenocompounds with anticancer and efflux pump inhibitory activity using preliminary computational methods]. Acta Pharm. Hung. 2018, 88, 67–74. [Google Scholar]
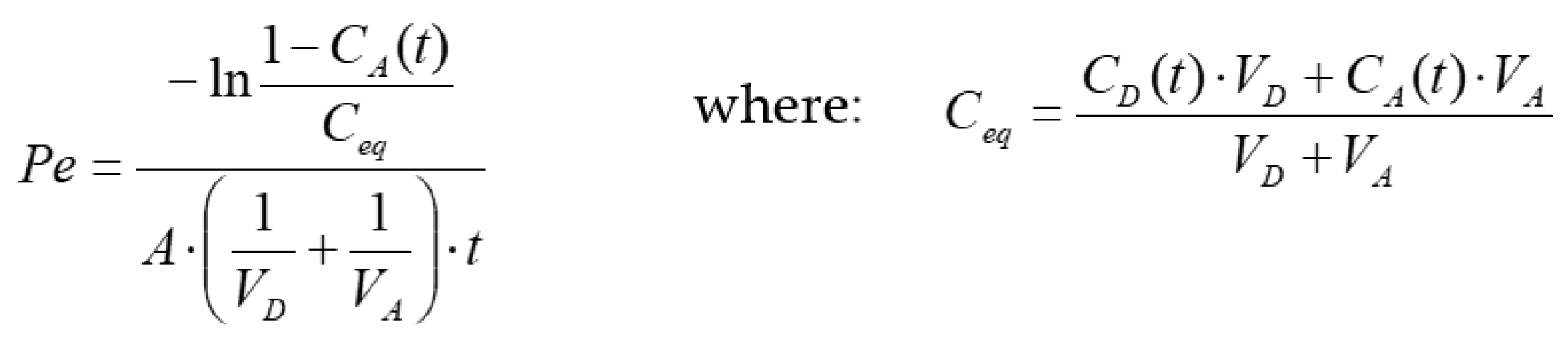
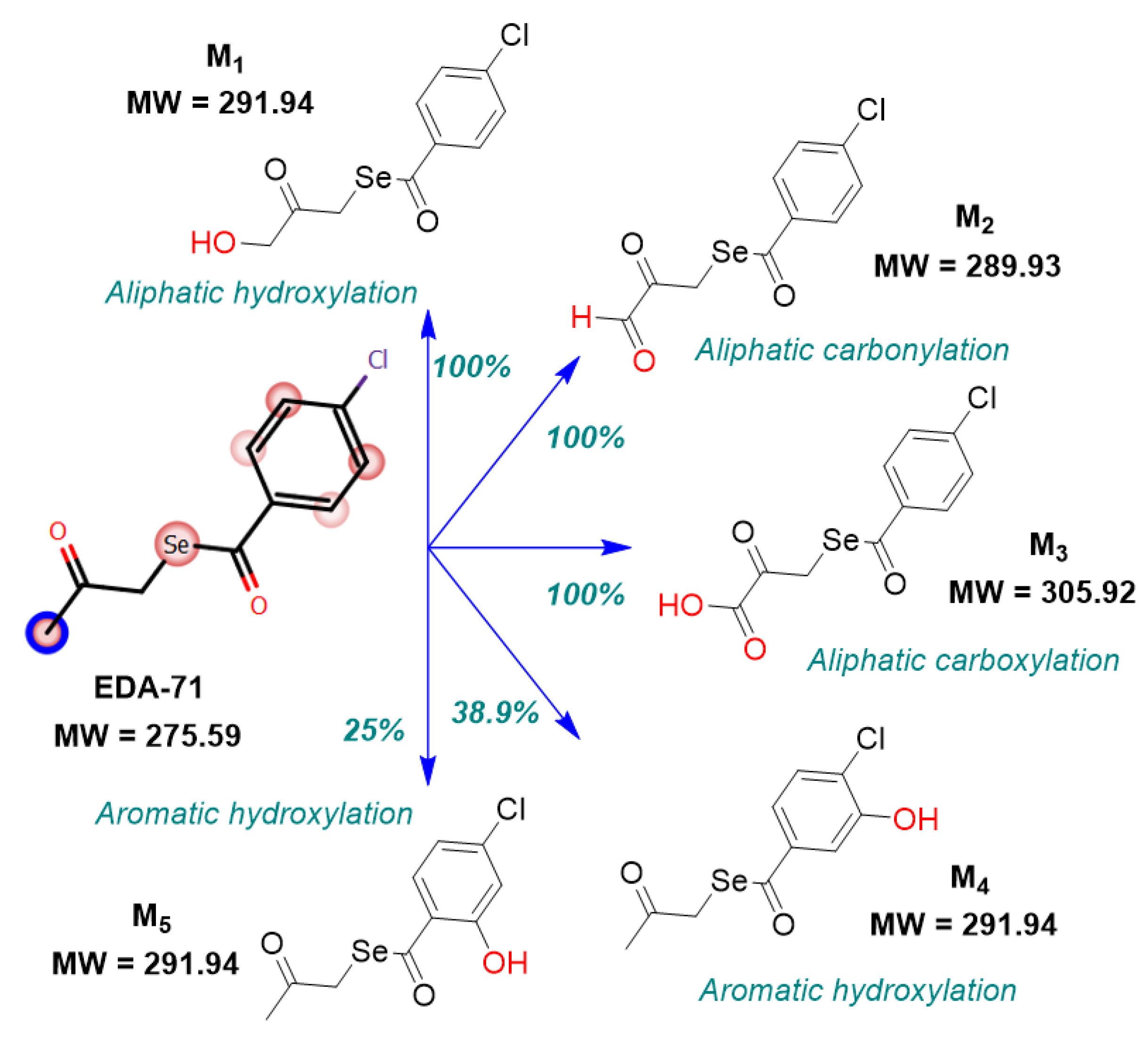

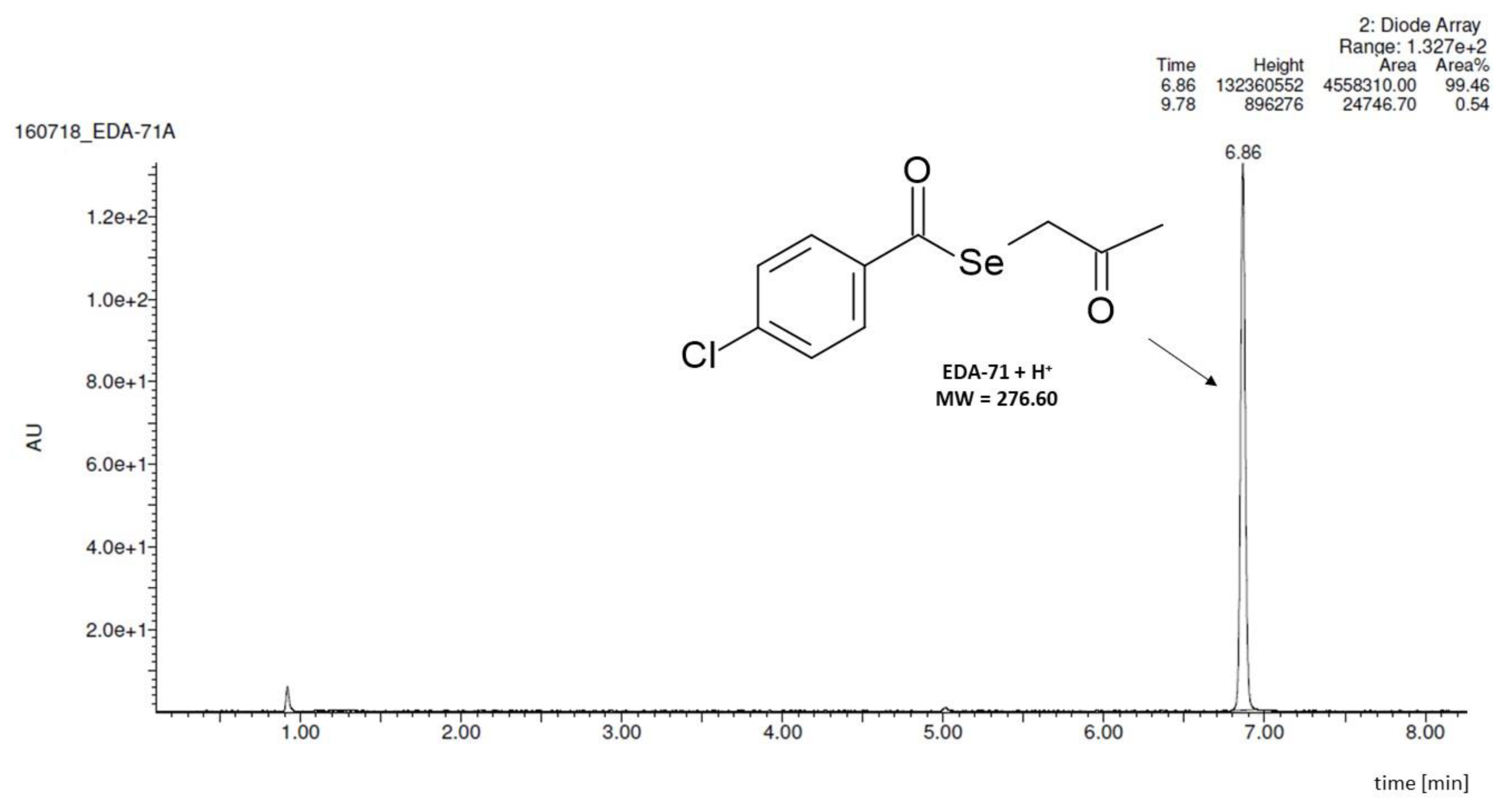
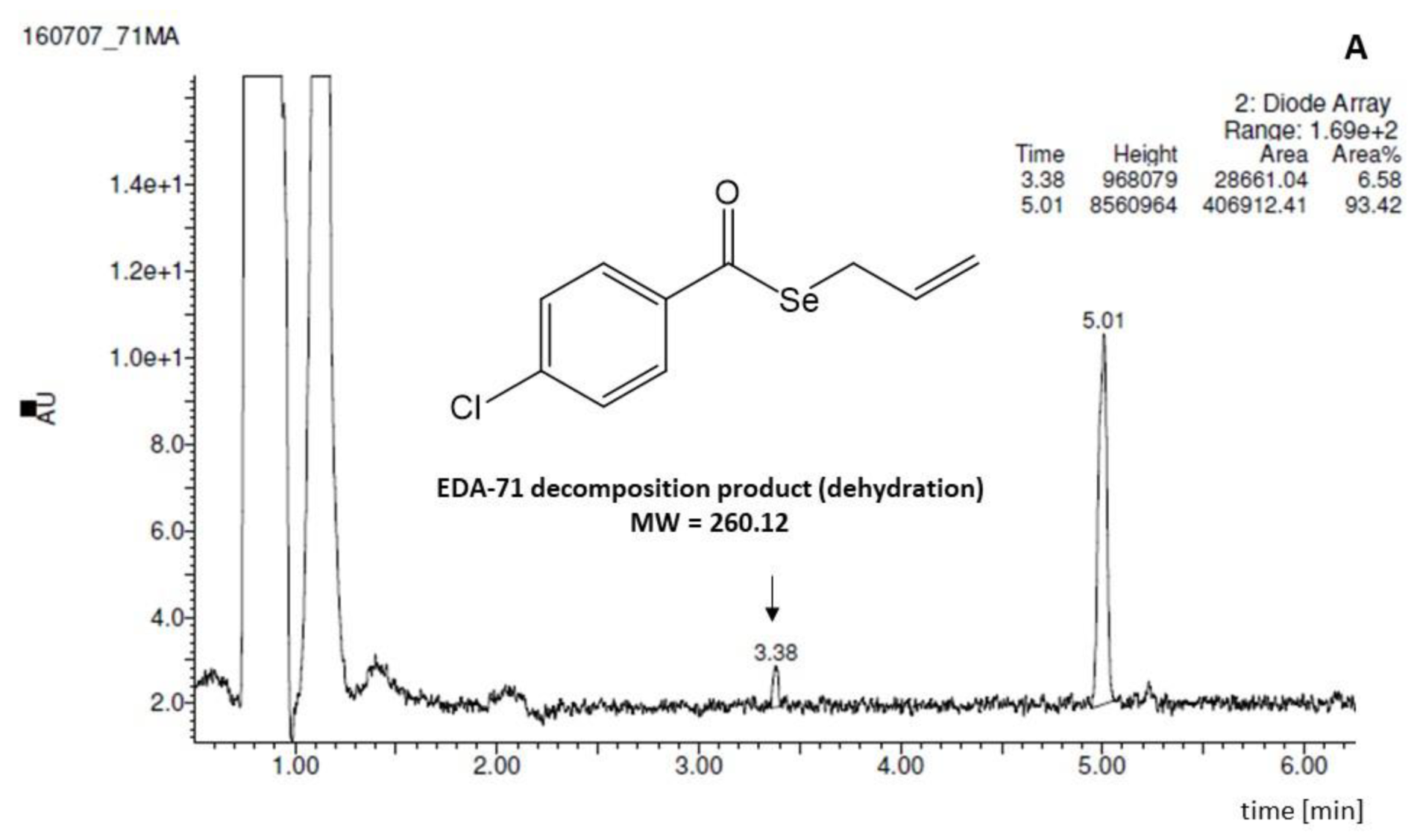
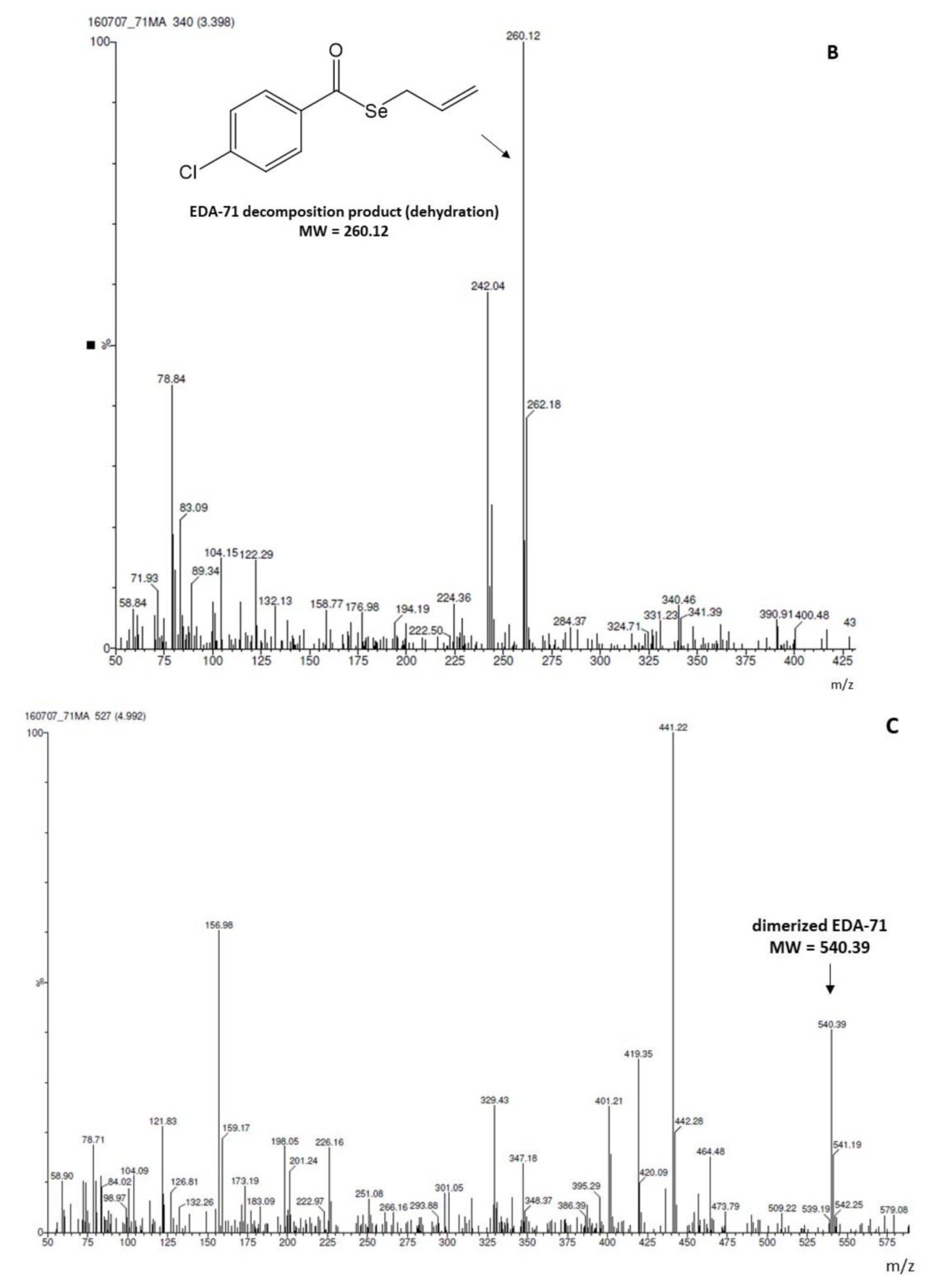
| Tested Compound | Experimental Aqueous Solubility | In Silico Calculated Aqueous Solubility | % That the In Silico Value Supposes in Respect to the Aqueous Experimental Solubility | |||||
|---|---|---|---|---|---|---|---|---|
| [g/L] | logSm | Chemic logSm | SwissA logSm 1 | OSIR logSm | Chemic | SwissA | OSIR | |
| EDA-26 | 0.380 | −2.96 | −4.02 | −5.20 | −5.06 | 8.71% | 0.58% | 0.79% |
| EDA-71 | 0.132 | −3.32 | −4.85 | −5.53 | −5.96 | 2.95% | 0.62% | 0.23% |
| EDA-73 | ~0.001 | ~−5.50 | −5.82 | −5.59 | −6.63 | 47.9% | 81.3% | 7.41% |
| EDA-74 | 0.019 | −4.26 | −5.10 | −6.03 | −5.97 | 14.5% | 1.70% | 1.95% |
| EDA-93 | 0.022 | −4.16 | −5.95 | −6.40 | −6.30 | 1.62% | 0.58% | 0.72% |
| EDA-109 | 0.028 | −4.06 | −3.53 | −6.13 | −6.16 | 339% | 0.85% | 0.79% |
| EDA-117 | 1.028 | −2.37 | −3.81 | −4.21 | −4.79 | 3.63% | 1.45% | 0.38% |
| EDA-119 | 0.137 | −3.37 | −2.82 | −5.88 | −5.41 | 355% | 0.31% | 0.91% |
| EDA-120 | 0.026 | −4.09 | −3.53 | −6.13 | −6.16 | 363% | 0.91% | 0.85% |
| EDA-122 | 0.031 | −4.02 | −3.76 | −6.35 | −6.28 | 182% | 0.47% | 0.55% |
| EDA-A6 | 0.758 | −2.44 | −4.29 | −4.91 | −5.68 | 1.41% | 0.34% | 0.06% |
| Compound | In Vitro Permeability [cm/s] (% Perm.) | Compound | In Vitro Permeability [cm/s] (% Perm.) | Reference | In Vitro Permeability [cm/s] (% Perm.) |
|---|---|---|---|---|---|
| EDA-58 | 2.220 × 10−6 (3.95%) | EDA-122 | 4.668 × 10−6 (8.20%) | Caffeine | 3.420 × 10−6 (6.05%) |
| EDA-71 | 3.858 × 10−6 (6.81%) | EDA-A6 | 3.438 × 10−6 (6.08%) | Norfloxacin | 0.9488 × 10−7 (1.70%) |
| EDA-119 | 3.271 × 10−6 (5.79%) |
| Compound | Mutagenicity at 1 µM Concentration | Mutagenicity at 10 µM Concentration | ||||||
|---|---|---|---|---|---|---|---|---|
| MI 1 | Fold Increase over Baseline | B 2 | SD | MI | Fold Increase over Baseline | B | SD | |
| MetSeAc | 0.90 | 0.75 | 0.37 | 2.65 | 0.96 | 0.74 | 0.47 | 1.53 |
| DOXO | 1.64 | 1.31 | 1.00 | 1.00 | 0.95 | 0.77 | 0.47 | 3.61 |
| EDA-109 | 0.96 | 0.55 | 0.07 | 0.58 | 0.67 | 0.39 | 0.003 C | 1.73 |
| EDA-119 | 0.56 | 0.54 | 0.24 | 0.58 | 0.96 | 0.92 | 0.90 | 2.52 |
| EDA-120 | 1.24 | 1.23 | 1.00 | 1.53 | 1.46 | 1.45 | 1.00 | 2.08 |
| EDA-122 | 0.84 | 0.63 | 0.14 | 3.61 | 1.46 | 1.08 | 0.94 | 2.89 |
| Compound | Mutagenicity at 1 µM Concentration | Mutagenicity at 10 µM Concentration | ||||||
|---|---|---|---|---|---|---|---|---|
| MI | Fold Increase over Baseline | B | SD | MI | Fold Increase over Baseline | B | SD | |
| MetSeAc | 0.90 | 0.75 | 0.37 | 2.65 | 0.96 | 0.74 | 0.47 | 1.53 |
| DOXO | 1.64 | 1.31 | 1.00 | 1.00 | 0.95 | 0.77 | 0.46 | 3.61 |
| EDA-53 | 1.82 | 1.10 | 1.00 | 12.33 | 1.72 | 1.04 | 0.98 | 11.67 |
| EDA-71 | 0.54 | 0.60 | 0.34 | 1.15 | 0.79 | 0.89 | 0.98 | 1.53 |
| EDA-73 | 1.28 | 0.84 | 0.72 | 0.58 | 0.79 | 0.52 | 0.04 | 4.04 |
| EDA-74 | 0.88 | 0.98 | 0.94 | 1.00 | 0.69 | 0.76 | 0.68 | 0.58 |
| Compound | Mutagenicity at 1 µM Concentration | Mutagenicity at 10 µM Concentration | ||||||
|---|---|---|---|---|---|---|---|---|
| MI | Fold Increase over Baseline | B | SD | MI | Fold Increase over Baseline | B | SD | |
| MetSeAc | 0.90 | 0.75 | 0.37 | 2.65 | 0.96 | 0.74 | 0.47 | 1.53 |
| DOXO | 1.64 | 1.31 | 1.00 | 1.00 | 0.95 | 0.77 | 0.46 | 3.61 |
| EDA-26 | 1.51 | 0.96 | 0.92 | 4.73 | 2.16 | 1.37 | 1.00 | 2.08 |
| EDA-46 | 1.56 | 1.65 | 1.00 | 3.00 | 0.94 | 1.00 | 0.88 | 1.15 |
| EDA-56 | 1.46 | 0.93 | 0.88 | 2.89 | 0.85 | 0.90 | 0.75 | 6.56 |
| EDA-58 | 0.57 | 0.67 | 0.80 | 1.00 | 1.51 | 1.78 | 1.00 | 2.52 |
| EDA-93 | 1.27 | 1.02 | 0.96 | 1.00 | 1.08 | 0.87 | 0.80 | 1.53 |
| EDA-117 | 0.66 | 0.49 | 0.36 | 0.58 | 0.57 | 0.42 | 0.18 | 1.00 |
| EDA-A6 | 1.23 | 1.08 | 0.94 | 4.04 | 0.38 | 0.33 | 0.001 C | 3.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marć, M.A.; Domínguez-Álvarez, E.; Latacz, G.; Doroz-Płonka, A.; Sanmartín, C.; Spengler, G.; Handzlik, J. Pharmaceutical and Safety Profile Evaluation of Novel Selenocompounds with Noteworthy Anticancer Activity. Pharmaceutics 2022, 14, 367. https://doi.org/10.3390/pharmaceutics14020367
Marć MA, Domínguez-Álvarez E, Latacz G, Doroz-Płonka A, Sanmartín C, Spengler G, Handzlik J. Pharmaceutical and Safety Profile Evaluation of Novel Selenocompounds with Noteworthy Anticancer Activity. Pharmaceutics. 2022; 14(2):367. https://doi.org/10.3390/pharmaceutics14020367
Chicago/Turabian StyleMarć, Małgorzata Anna, Enrique Domínguez-Álvarez, Gniewomir Latacz, Agata Doroz-Płonka, Carmen Sanmartín, Gabriella Spengler, and Jadwiga Handzlik. 2022. "Pharmaceutical and Safety Profile Evaluation of Novel Selenocompounds with Noteworthy Anticancer Activity" Pharmaceutics 14, no. 2: 367. https://doi.org/10.3390/pharmaceutics14020367