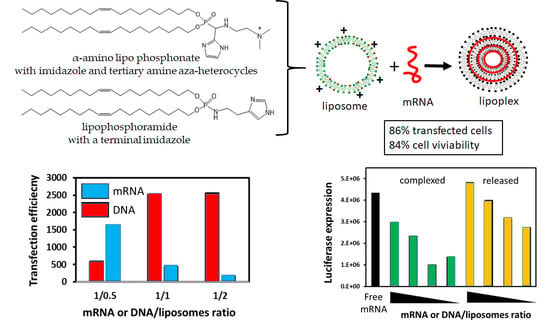
mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Transcribed mRNA
2.2. Plasmid DNA (pDNA)
2.3. Cells and Cell Culture
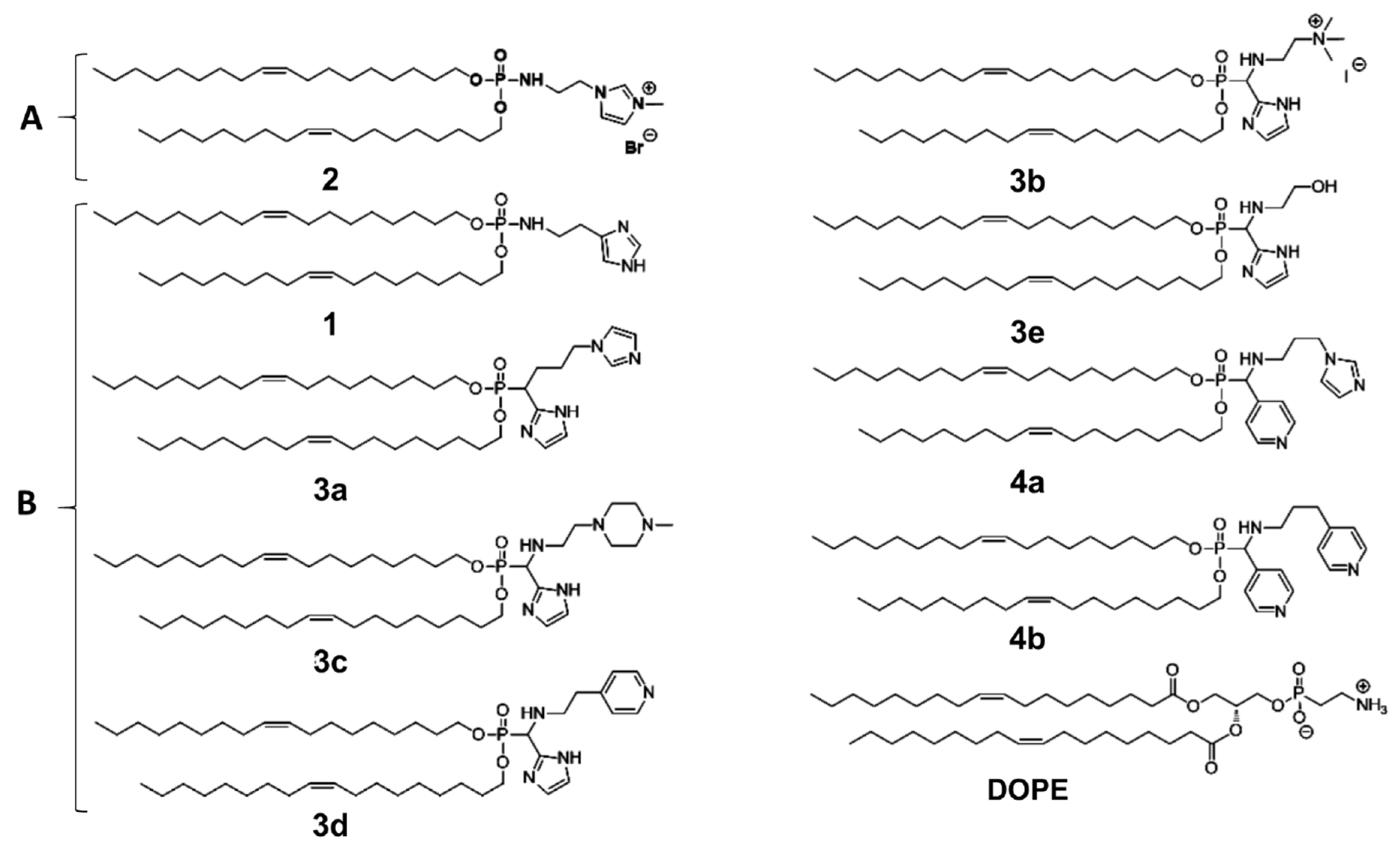
2.4. Liposomes
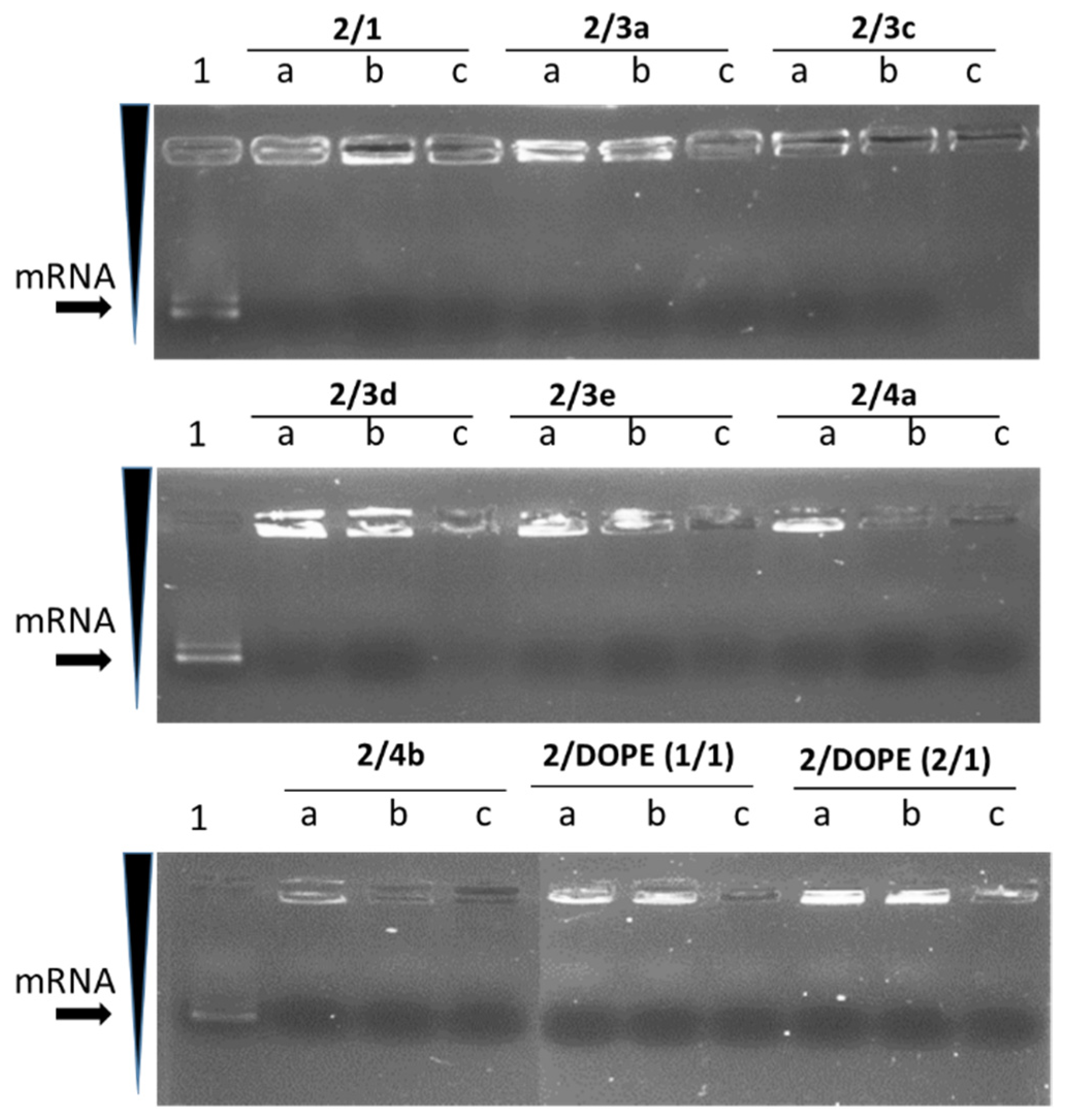
2.5. Lipoplexes (LX) Preparation
2.6. Size Distribution and ζ Potential Measurements
2.7. Membrane Fusion Study
2.8. Transfection
2.9. Cytotoxicity Assay
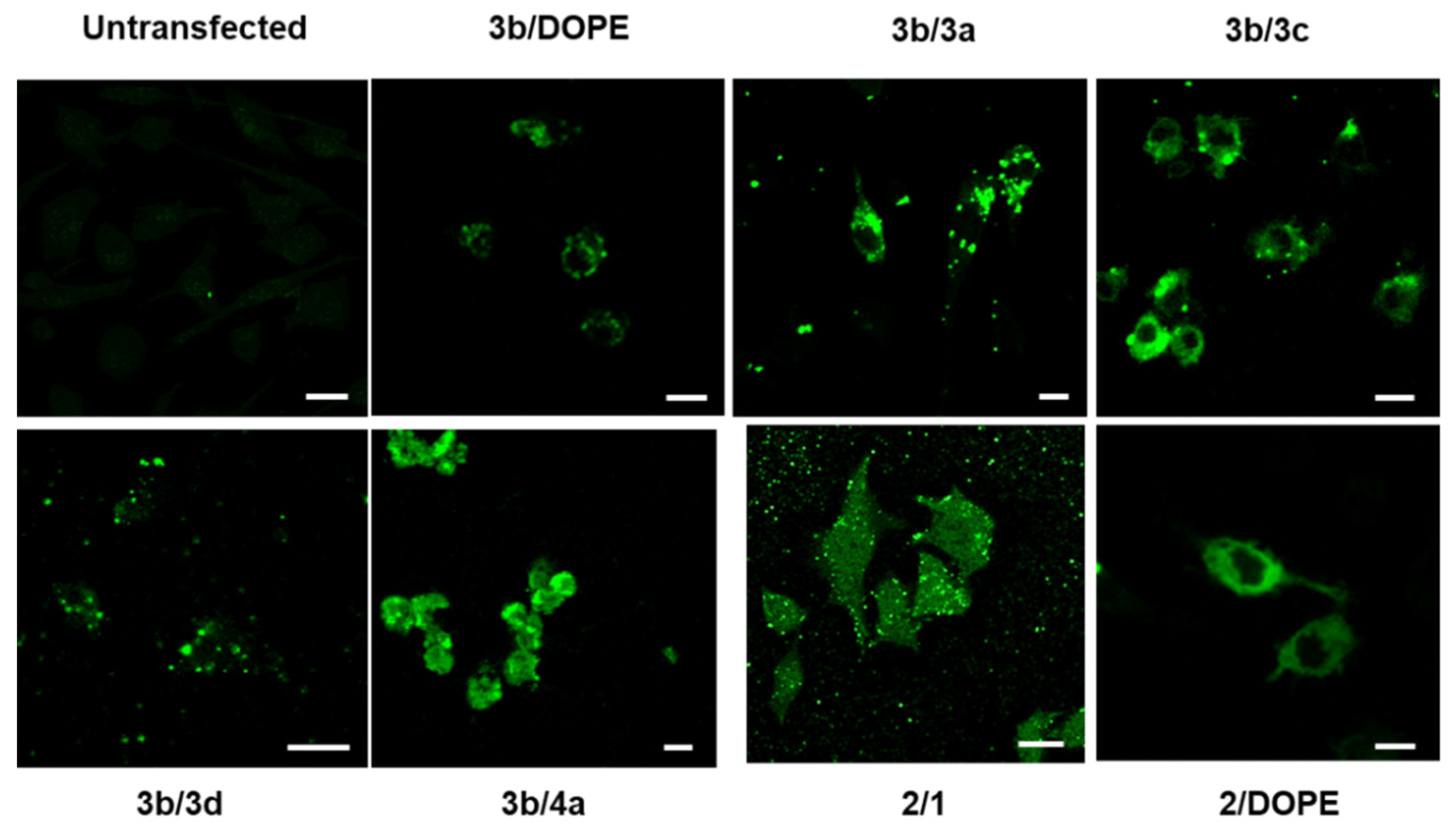
2.10. Laser Scanning Confocal Microscopy
2.11. Reticulocyte Assay
2.12. Circular Dichroism Spectroscopy (CD)
3. Results and Discussion
3.1. Liposomes Physicochemical Properties
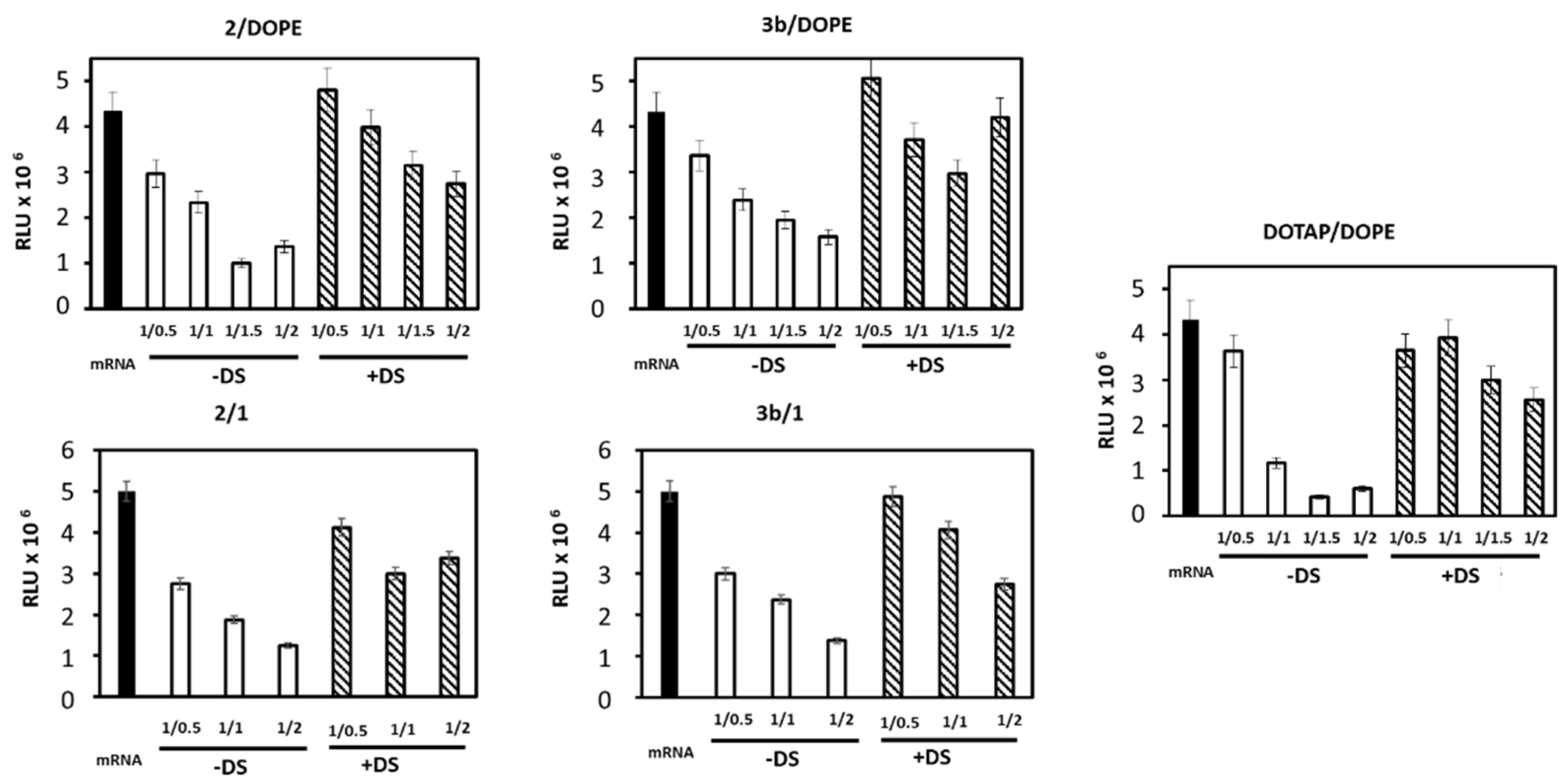
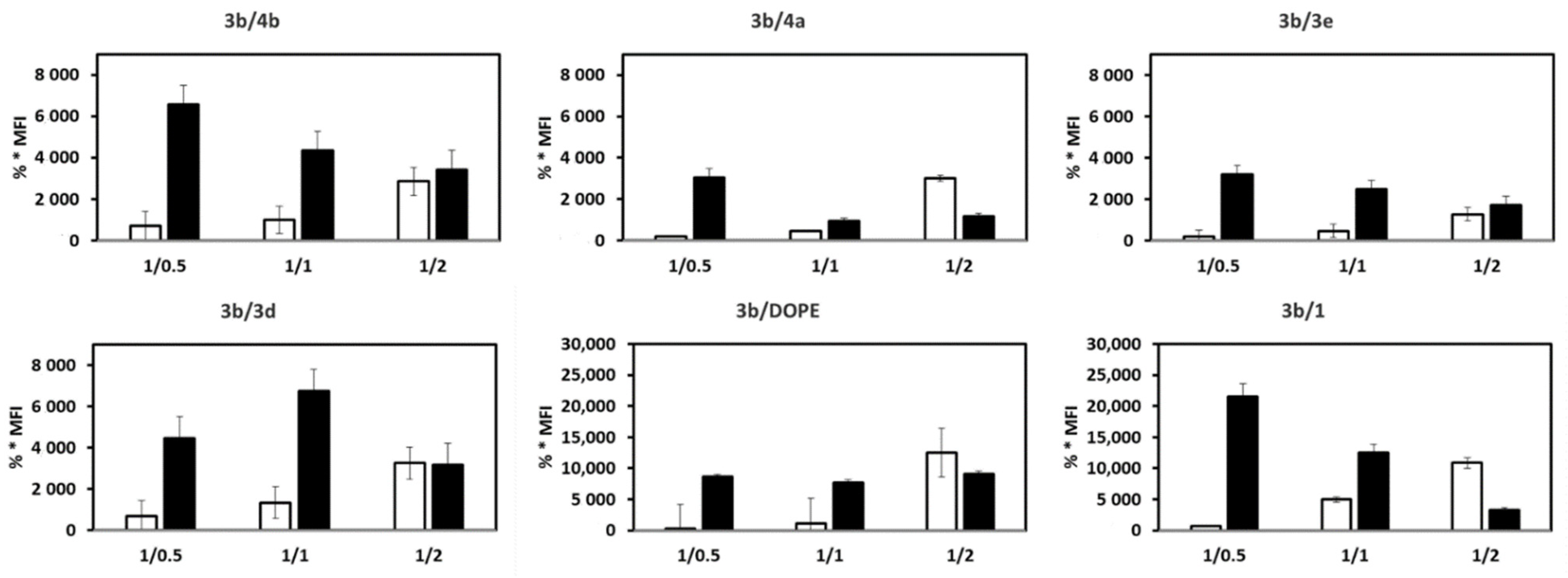
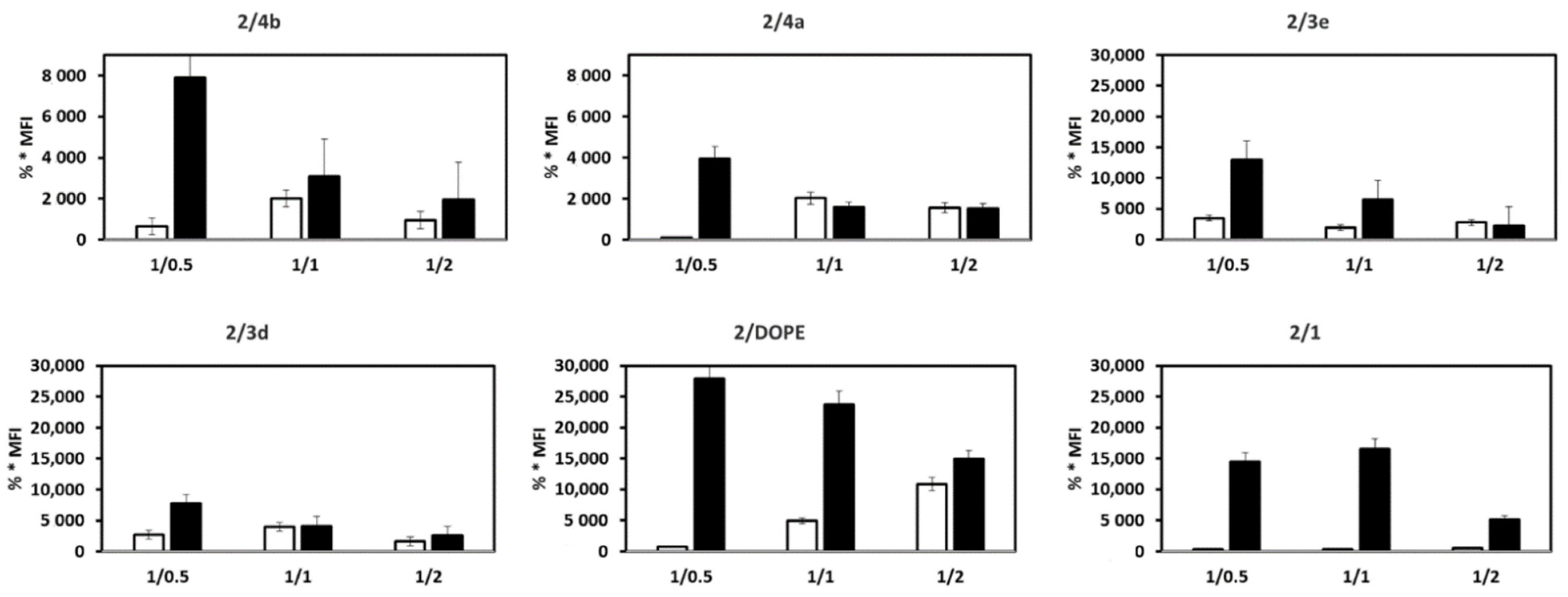
3.2. mRNA Transfection
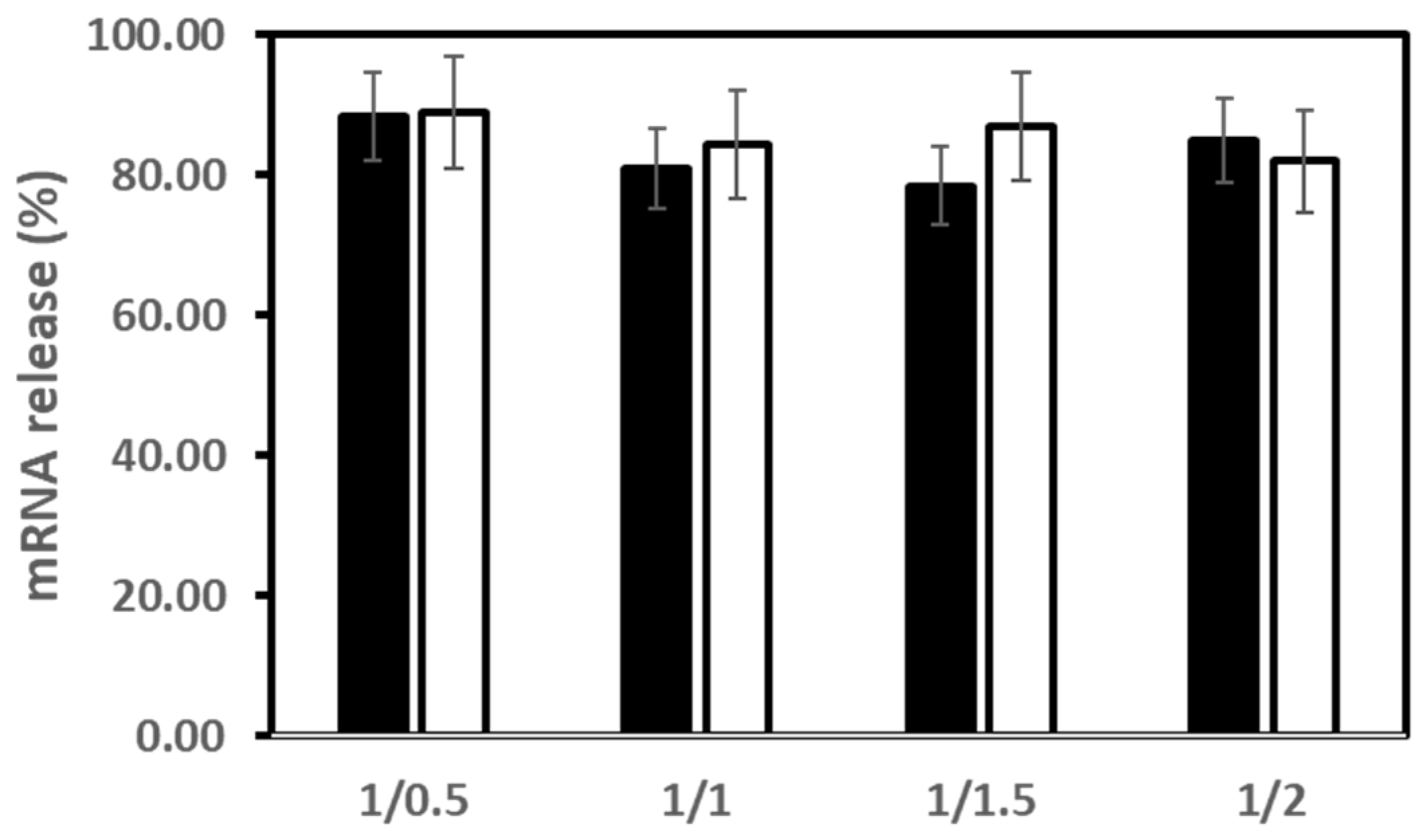
3.3. mRNA Release from LX
3.4. Translation Capacity Post-Release mRNA
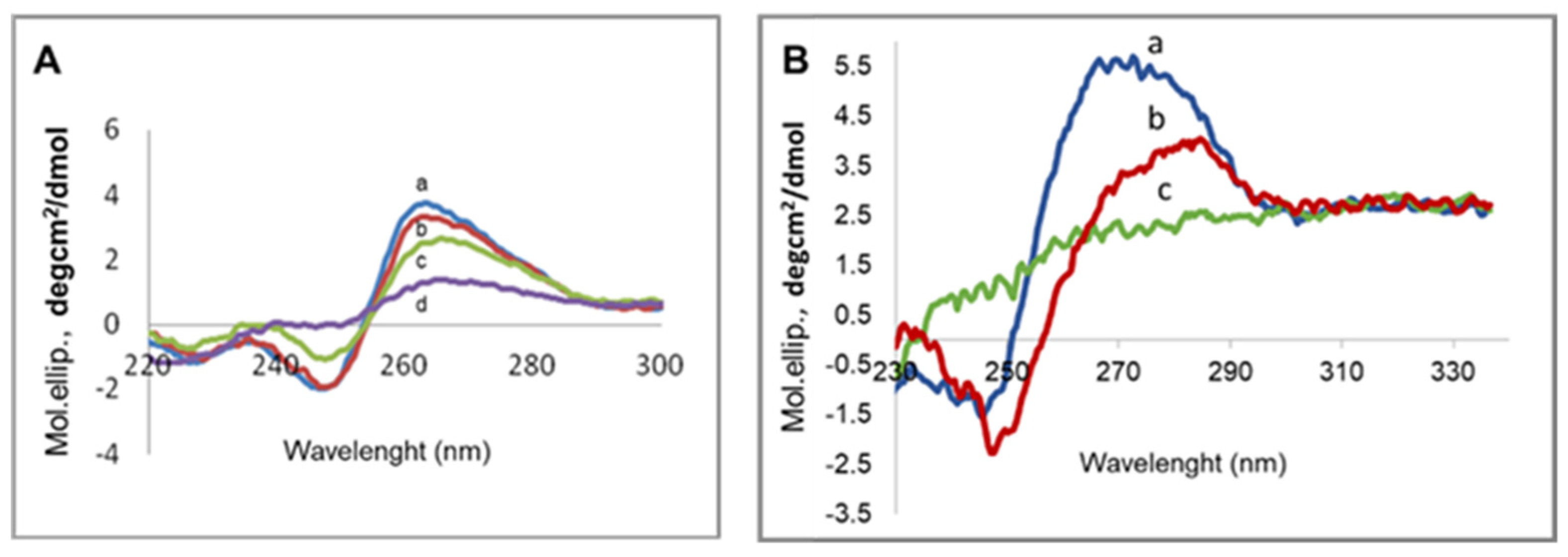
3.5. mRNA Conformation
3.6. mRNA Transfection versus DNA Transfection
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Siewert, C.D.; Haas, H.; Cornet, V.; Nogueira, S.S.; Nawroth, T.; Uebbing, L.; Ziller, A.; Al-Gousous, J.; Radulescu, A.; Schroer, M.A.; et al. Hybrid Biopolymer and Lipid Nanoparticles with Improved Transfection Efficacy for mRNA. Cells 2020, 9, 2034. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. RNA-Based COVID-19 Vaccine BNT162b2 Selected for a Pivotal Efficacy Study. medRxiv 2020. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Ballesteros-Briones, M.C.; Silva-Pilipich, N.; Herrador-Canete, G.; Vanrell, L.; Smerdou, C. A new generation of vaccines based on alphavirus self-amplifying RNA. Curr. Opin. Virol. 2020, 44, 145–153. [Google Scholar] [CrossRef]
- Bloom, K.; van den Berg, F.; Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020, 28, 117–129. [Google Scholar] [CrossRef]
- Chow, M.Y.; Chang, R.Y.K.; Chan, H.K. Inhalation delivery technology for genome-editing of respiratory diseases. Adv. Drug Deliv. Rev. 2020, 168, 217–228. [Google Scholar] [CrossRef]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef]
- Li, J.; Roise, J.J.; He, M.; Das, R.; Murthy, N. Non-viral strategies for delivering genome editing enzymes. Adv. Drug Deliv. Rev. 2020, 168, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Luisi, K.; Morabito, K.M.; Burgomaster, K.E.; Sharma, M.; Kong, W.P.; Foreman, B.M.; Patel, S.; Fisher, B.; Aleshnick, M.A.; Laliberte, J.; et al. Development of a potent Zika virus vaccine using self-amplifying messenger RNA. Sci. Adv. 2020, 6, eaba5068. [Google Scholar] [CrossRef] [PubMed]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Lower, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3’ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, E.; MacDonald, K.D.; Slaughter, K.; McKinney, M.; Patel, S.; Sun, C.; Sahay, G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018, 26, 2034–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Lower, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrors, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef]
- Trepotec, Z.; Lichtenegger, E.; Plank, C.; Aneja, M.K.; Rudolph, C. Delivery of mRNA Therapeutics for the Treatment of Hepatic Diseases. Mol. Ther. 2019, 27, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Xiangjun Tang, X.; Zhang, S.; Fu, R.; Zhang, L.; Huang, K.; Peng, H.; Dai, L.; Chen, Q. Therapeutic Prospects of mRNA-Based Gene Therapy for Glioblastoma. Front. Oncol. 2019, 1208. [Google Scholar] [CrossRef]
- Zuhorn, I.S.; Oberle, V.; Visser, W.H.; Engberts, J.B.; Bakowsky, U.; Polushkin, E.; Hoekstra, D. Phase behavior of cationic amphiphiles and their mixtures with helper lipid influences lipoplex shape, DNA translocation, and transfection efficiency. Biophys. J. 2002, 83, 2096–2108. [Google Scholar] [CrossRef] [Green Version]
- Dal-Maso, A.D.; Dellacasagrande, J.; Legendre, F.; Tiraby, G.; Blonski, C.; Hoffmann, P. Synthesis and evaluation of new phosphonolipid compounds for gene delivery. Eur. J. Med. Chem. 2008, 43, 1758–1766. [Google Scholar] [CrossRef]
- Du, Z.; Munye, M.M.; Tagalakis, A.D.; Manunta, M.D.; Hart, S.L. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci. Rep. 2014, 4, 7107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta 2013, 1828, 412–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, J.P.; Aytar, B.S.; Kondo, Y.; Lynn, D.M.; Abbott, N.L. Incorporation of DOPE into Lipoplexes formed from a Ferrocenyl Lipid leads to Inverse Hexagonal Nanostructures that allow Redox-Based Control of Transfection in High Serum. Soft Matter 2012, 8, 2608–2619. [Google Scholar] [CrossRef] [PubMed]
- Safinya, C.R. Structures of lipid-DNA complexes: Supramolecular assembly and gene delivery. Curr. Opin. Struct. Biol. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Mevel, M.; Breuzard, G.; Yaouanc, J.J.; Clement, J.C.; Lehn, P.; Pichon, C.; Jaffres, P.A.; Midoux, P. Synthesis and transfection activity of new cationic phosphoramidate lipids: High efficiency of an imidazolium derivative. ChemBioChem 2008, 9, 1462–1471. [Google Scholar] [CrossRef]
- Mevel, M.; Neveu, C.; Goncalves, C.; Yaouanc, J.J.; Pichon, C.; Jaffres, P.A.; Midoux, P. Novel neutral imidazole-lipophosphoramides for transfection assays. Chem. Commun. 2008, 27, 3124–3126. [Google Scholar] [CrossRef] [Green Version]
- Midoux, P.; Pichon, C.; Yaouanc, J.J.; Jaffres, P.A. Chemical vectors for gene delivery: A current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009, 157, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, C.; Berchel, M.; Gosselin, M.P.; Malard, V.; Cheradame, H.; Jaffres, P.A.; Guegan, P.; Pichon, C.; Midoux, P. Lipopolyplexes comprising imidazole/imidazolium lipophosphoramidate, histidinylated polyethyleneimine and siRNA as efficient formulation for siRNA transfection. Int. J. Pharm. 2014, 460, 264–272. [Google Scholar] [CrossRef]
- Perche, F.; Gosset, D.; Mevel, M.; Miramon, M.L.; Yaouanc, J.J.; Pichon, C.; Benvegnu, T.; Jaffres, P.A.; Midoux, P. Selective gene delivery in dendritic cells with mannosylated and histidylated lipopolyplexes. J. Drug. Target. 2011, 19, 315–325. [Google Scholar] [CrossRef]
- Le Moignic, A.; Malard, V.; Benvegnu, T.; Lemiegre, L.; Berchel, M.; Jaffres, P.A.; Baillou, C.; Delost, M.; Macedo, R.; Rochefort, J.; et al. Preclinical evaluation of mRNA trimannosylated lipopolyplexes as therapeutic cancer vaccines targeting dendritic cells. J. Control. Release 2018, 278, 110–121. [Google Scholar] [CrossRef]
- Perche, F.; Benvegnu, T.; Berchel, M.; Lebegue, L.; Pichon, C.; Jaffres, P.A.; Midoux, P. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine 2011, 7, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Van der Jeught, K.; De Koker, S.; Bialkowski, L.; Heirman, C.; Tjok Joe, P.; Perche, F.; Maenhout, S.; Bevers, S.; Broos, K.; Deswarte, K.; et al. Dendritic Cell Targeting mRNA Lipopolyplexes Combine Strong Antitumor T-Cell Immunity with Improved Inflammatory Safety. ACS Nano 2018, 12, 9815–9829. [Google Scholar] [CrossRef] [PubMed]
- Berchel, M.; Akhter, S.; Berthe, W.; Goncalves, C.; Dubuisson, M.; Pichon, C.; Jaffres, P.A.; Midoux, P. Synthesis of alpha-amino-lipophosphonates as cationic lipids or co-lipids for DNA transfection in dendritic cells. J. Mater. Chem. B 2017, 5, 6869–6881. [Google Scholar] [CrossRef] [PubMed]
- Mockey, M.; Goncalves, C.; Dupuy, F.P.; Lemoine, F.M.; Pichon, C.; Midoux, P. mRNA transfection of dendritic cells: Synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006, 340, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Reznikoff, G.; Dranoff, G.; Rock, K.L. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 1997, 158, 2723–2730. [Google Scholar]
- Kumar, V.V.; Pichon, C.; Refregiers, M.; Guerin, B.; Midoux, P.; Chaudhuri, A. Single histidine residue in head-group region is sufficient to impart remarkable gene transfection properties to cationic lipids: Evidence for histidine-mediated membrane fusion at acidic pH. Gene Ther. 2003, 10, 1206–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gratzer, W.B.; Hill, L.R.; Owen, R.J. Circular dichroism of DNA. Eur. J. Biochem. 1970, 15, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Suga, K.; Tanabe, T.; Tomita, H.; Shimanouchi, T.; Umakoshi, H. Conformational change of single-stranded RNAs induced by liposome binding. Nucleic Acids Res. 2011, 39, 8891–8900. [Google Scholar] [CrossRef] [Green Version]
- Voet, D.; Gratzer, W.; Cox, R.; Doty, P. Absorption spectra of nucleotides, polynucleotides, and nucleic acids in the far ultraviolet. Biopolymers 1963, 1, 193–208. [Google Scholar] [CrossRef]
- Suga, K.; Tanabe, T.; Umakoshi, H. Heterogeneous cationic liposomes modified with 3beta-{N-[(N’,N’-dimethylamino)ethyl]carbamoyl}cholesterol can induce partial conformational changes in messenger RNA and regulate translation in an Escherichia coli cell-free translation system. Langmuir 2013, 29, 1899–1907. [Google Scholar] [CrossRef]
- Ciani, L.; Casini, A.; Gabbiani, C.; Ristori, S.; Messori, L.; Martini, G. DOTAP/DOPE and DC-Chol/DOPE lipoplexes for gene delivery studied by circular dichroism and other biophysical techniques. Biophys. Chem. 2007, 127, 213–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Co-Lipids | Mol. Ratio £ | Size (nm) | PDI * | ζ Potential (mV) |
|---|---|---|---|---|
| 3a | 1/1 | 235 ± 2 | 0.3 | 49.4 ± 0.6 |
| 3c | 1/1 | 102 ± 1 | 0.2 | 56 ± 0.5 |
| 3d | 1/1 | 143 ± 3 | 0.4 | 71 ± 1 |
| 3e | 1/1 | 85 ± 1 | 0.1 | 73 ± 0.1 |
| 4a | 1/1 | 140 ± 2 | 0.1 | 72 ± 1 |
| 4b | 1/1 | 139 ± 2 | 0.5 | 67 ± 0.7 |
| 1 | 1/1 | 165 ± 2 | 0.4 | 70 ± 0.8 |
| DOPE 1/1 | 1/1 | 120 ± 1 | 0.1 | 79 ± 0.6 |
| DOPE 2/1 | 2/1 | 116 ± 2 | 0.3 | 84 ± 1.1 |
| Co-Lipids | Mol. Ratio £ | Size (nm) | PDI * | ζ Potential (mV) |
|---|---|---|---|---|
| 3a | 1/1 | 101 ± 1 | 0.5 | 76 ± 0.7 |
| 3c | 1/1 | 106 ± 1 | 0.5 | 71 ± 0.5 |
| 3d | 1/1 | 112 ± 2 | 0.3 | 71 ± 1.0 |
| 3e | 1/1 | 117 ± 2 | 0.2 | 71 ± 0.9 |
| 4a | 1/1 | 113 ± 1 | 0.2 | 73 ± 1.2 |
| 4b | 1/1 | 163 ± 2 | 0.3 | 53 ± 1.0 |
| 1 | 1/1 | 96 ± 1 | 0.3 | 74 ± 0.5 |
| DOPE 1/1 | 1/1 | 163 ± 1 | 0.1 | 67 ± 0.6 |
| DOPE 2/1 | 2/1 | 122 ± 2 | 0.1 | 81 ± 1.1 |
| 3b-Based Liposomes | 2-Based Liposomes | |||
|---|---|---|---|---|
| Co-Lipids | pH 7.4 | pH 5.5 | pH 7.4 | pH 5.5 |
| 3a | 20 ± 1 | 79 ± 7 | 0 | 0 |
| 3c | 0 | 41 ± 4 | 0 | 4.5 ± 0.5 |
| 3d | 0 | 2.6 | 0 | 16 ± 1 |
| 3e | 31 ± 2 | 21 ± 2 | 31 ± 3 | 10 ± 1 |
| 4a | 0 | 37 ± 3 | 0 | 0 |
| 4b | 8 ± 1 | 0 | 0 | 11 ± 1 |
| 1 | 34 ± 3 | 71 ± 6 | 21 ± 2 | 97 ± 9 |
| DOPE 1/1 | 1.2 | 19 ± 2 | 12 ± 1 | 58 ± 6 |
| DOPE 2/1 | 0 | 58 ± 5 | 0 | 99 ± 3 |
| Co-Lipids | 3b-Based LXs | 2-Based LXs | |||
|---|---|---|---|---|---|
| mRNA/L | 1/0.5 | 1/1 | 1/0.5 | 1/1 | |
| 3a | 94 ± 7 | 85 ± 7 | 94 ± 7 | 95 ± 7 | |
| 3c | 93 ± 7 | 94 ± 8 | 85 ± 7 | 42 ± 3 | |
| 3d | 88 ± 7 | 72 ± 5 | 73 ± 6 | 69 ± 5 | |
| 3e | 95 ± 8 | 70 ± 5 | 83 ± 7 | 84 ± 7 | |
| 4a | 76 ± 6 | 79 ± 6 | 85 ± 7 | 100 ± 8 | |
| 4b | 93 ± 7 | 84 ± 7 | 73 ± 6 | 57 ± 4 | |
| 1 | 83 ± 6 | 100 ± 8 | 69 ± 5 | 30 ± 2 | |
| DOPE1/1 | 80 ± 6 | 87 ± 7 | 92 ± 7 | 90 ± 7 | |
| DOPE2/1 | 84 ± 7 | 87 ± 7 | 97 ± 8 | 89 ± 7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhter, S.; Berchel, M.; Jaffrès, P.-A.; Midoux, P.; Pichon, C. mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation. Pharmaceutics 2022, 14, 581. https://doi.org/10.3390/pharmaceutics14030581
Akhter S, Berchel M, Jaffrès P-A, Midoux P, Pichon C. mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation. Pharmaceutics. 2022; 14(3):581. https://doi.org/10.3390/pharmaceutics14030581
Chicago/Turabian StyleAkhter, Sohail, Mathieu Berchel, Paul-Alain Jaffrès, Patrick Midoux, and Chantal Pichon. 2022. "mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation" Pharmaceutics 14, no. 3: 581. https://doi.org/10.3390/pharmaceutics14030581