Development and Characterization of Pharmaceutical Systems Containing Rifampicin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Phase Solubility Studies
2.3. Molecular Modeling
2.3.1. Modeling of Compounds
2.3.2. Molecular Docking Studies
2.3.3. Molecular Dynamics Simulations
2.3.4. Selection of Target Temperature for MD Simulations
2.4. Proton Nuclear Magnetic Resonance Spectroscopy
2.5. Preparation of Inclusion Complexes in Solid State
2.6. Fourier Transform-Infrared Spectroscopy and Powder X-ray Diffraction
2.7. Scanning Electron Microscopy
2.8. Dissolution Studies
2.9. Microbiological Studies
2.9.1. Bacterial Strain and Growth Conditions
2.9.2. Determination of the Minimum Inhibitory Concentration of RIF
2.9.3. In Vitro Antimicrobial Study
2.9.4. Evaluation of the Metabolic Activity of the Biofilm
Biofilm Formation and Treatment
XTT Assay
2.9.5. Biofilm Analysis by Scanning Electron Microscopy
Biofilm Preparation
Sample Preparation for SEM
2.10. Antileishmanial Activity
2.10.1. Promastigotes Culture
2.10.2. Antipromastigote Assay
Antipromastigote Assay
2.11. Stability Studies
2.11.1. Chromatographic Conditions
2.11.2. Degradation Studies
2.11.3. Validation Method
Linearity
Accuracy
Precision
Detection and Quantification Limits
Stability Evaluation
3. Results and Discussion
3.1. Phase Solubility Studies
3.2. Molecular Modeling of Binary Complexes
3.2.1. Molecular Modeling of Binary Complexes
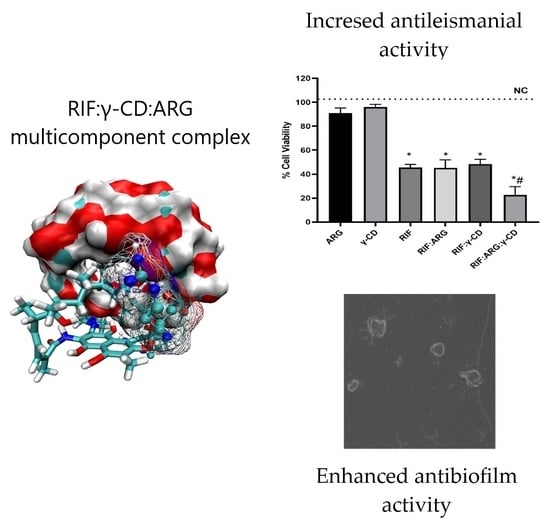
3.2.2. Molecular Modeling of Multicomponent Complexes
3.2.3. Interaction between RIF and ARG
3.3. Proton Nuclear Magnetic Resonance Spectroscopy
3.4. Characterization of Systems in the Solid State by Fourier Transform-Infrared Spectroscopy, Powder X-ray Diffraction, Thermal Analysis and Scanning Electron Microscopy
3.5. Dissolution Studies
3.6. Microbiological Studies
3.6.1. In Vitro Antimicrobial Study
3.6.2. Evaluation of the Metabolic Activity of the Biofilm
3.6.3. Biofilm Analysis by SEM
3.7. Antileishmanial Activity
3.8. Stability Studies
3.8.1. Method Validation

3.8.2. Stability Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rohr, J.R.; Barrett, C.B.; Civitello, D.J.; Craft, M.E.; Delius, B.; DeLeo, G.A.; Hudson, P.J.; Jouanard, N.; Nguyen, K.H.; Ostfeld, R.S.; et al. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019, 2, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Sartini, S.; Permana, A.D.; Mitra, S.; Tareq, A.M.; Salim, E.; Ahmad, I.; Harapan, H.; Emran, T.B.; Nainu, F. Current State and Promising Opportunities on Pharmaceutical Approaches in the Treatment of Polymicrobial Diseases. Pathogens 2021, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Aiassa, V.; Zoppi, A.; Albesa, I.; Longhi, M.R. Inclusion complexes of chloramphenicol with β-cyclodextrin and aminoacids as a way to increase drug solubility and modulate ROS production. Carbohydr. Polym. 2015, 121, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Aiassa, V.; Zoppi, A.; Becerra, M.C.; Albesa, I.; Longhi, M.R. Enhanced inhibition of bacterial biofilm formation and reduced leukocyte toxicity by chloramphenicol: β-cyclodextrin: N-acetylcysteine complex. Carbohydr. Polym. 2016, 152, 672–678. [Google Scholar] [CrossRef]
- Cerutti, J.P.; Aiassa, V.; Fernandez, M.A.; Longhi, M.R.; Quevedo, M.A.; Zoppi, A. Structural, physicochemical and biological characterization of chloramphenicol multicomponent complexes. J. Mol. Liq. 2021, 331, 115761. [Google Scholar] [CrossRef]
- Zoppi, A.; Buhlman, N.; Cerutti, J.P.; Longhi, M.R.; Aiassa, V. Influence of proline and β-Cyclodextrin in ketoconazole physicochemical and microbiological performance. J. Mol. Struct. 2019, 1176, 470–477. [Google Scholar] [CrossRef]
- Zoppi, A.; Bartolilla, A.; Longhi, M.R.; Aiassa, V. Simultaneous improvement of ketoconazole solubility, antifungal and antibiofilm activity by multicomponent complexation. Ther. Deliv. 2020, 11, 701–712. [Google Scholar] [CrossRef]
- Morin-Crini, N.; Fourmentin, S.; Fenyvesi, É.; Lichtfouse, E.; Torri, G.; Fourmentin, M.; Crini, G. 130 years of cyclodextrin discovery for health, food, agriculture, and the industry: A review. Environ. Chem. Lett. 2021, 19, 2581–2617. [Google Scholar] [CrossRef]
- Jansook, P.; Ogawa, N.; Loftsson, T. Cyclodextrins: Structure, physicochemical properties and pharmaceutical applications. Int. J. Pharm. 2018, 535, 272–284. [Google Scholar] [CrossRef]
- Aiassa, V.; Garnero, C.; Longhi, M.R.; Zoppi, A. Cyclodextrin Multicomponent Complexes: Pharmaceutical Applications. Pharmaceutics 2021, 13, 1099. [Google Scholar] [CrossRef]
- Neuber, H. Leishmaniasis. J. Dtsch. Dermatol. Ges. 2008, 9, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Von Stebut, E. Leishmaniasis. JDDG J. Dtsch. Dermatol. Ges. 2015, 13, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Jolivet-Gougeon, A.; Bonnaure-Mallet, M. Biofilms as a mechanism of bacterial resistance. Drug Discov. Today Technol. 2014, 11, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Chadha, R.; Saini, A.; Gupta, S.; Arora, P.; Thakur, D.; Jain, D.V.S. Encapsulation of rifampicin by natural and modified β-cyclodextrins: Characterization and thermodynamic parameters. J. Incl. Phenom. Macrocycl. Chem. 2010, 67, 109–116. [Google Scholar] [CrossRef]
- Anjani, Q.K.; Domínguez-Robles, J.; Utomo, E.; Font, M.; Martínez-Ohárriz, M.C.; Permana, A.D.; Cárcamo-Martínez, Á.; Larrañeta, E.; Donnelly, R.F. Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization. Pharmaceuticals 2021, 15, 20. [Google Scholar] [CrossRef]
- Dan Córdoba, A.V.; Aiassa, V.; Longhi, M.R.; Quevedo, M.A.; Zoppi, A. Improved Activity of Rifampicin Against Biofilms of Staphylococcus aureus by Multicomponent Complexation. AAPS PharmSciTech 2020, 21, 163. [Google Scholar] [CrossRef]
- Higuchi, T.; Connors, K.A. Phase-solubility techniques. Adv. Anal. Chem. Instr. 1965, 4, 117–212. [Google Scholar]
- Available online: https://www.ccdc.cam.ac.uk/ (accessed on 5 November 2022).
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. Glycam06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://chemaxon.com/marvin (accessed on 5 November 2022).
- He, X.; Man, V.H.; Yang, W.; Lee, T.S.; Wang, J. A fast and high-quality charge model for the next generation general amber force field. J. Chem. Phys. 2020, 153, 114502. [Google Scholar] [CrossRef]
- Santos-Martins, D.; Solis-Vasquez, L.; Tillack, A.F.; Sanner, M.F.; Koch, A.; Forli, S. Accelerating autodock4 with gpus and gradient-based local search. J. Chem. Theory Comput. 2021, 17, 1060–1073. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using autodock for ligand-receptor docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R., III; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. Mmpbsa. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Case, D.; Aktulga, H.; Belfon, K.; Ben-Shalom, I.; Berryman, J.; Brozell, S.; Cerutti, D.; Cheatham, T., III; Cisneros, G.; Cruzeiro, V.; et al. Amber22. 2022. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and dynamics of the tip3p, spc, and spc/e water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.; Melo, M.N.; Seyler, S.L.; Domanski, J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. Mdanalysis: A python package for the rapid analysis of molecular dynamics simulations. In Proceedings of the 15th Python in Science Conference, SciPy, Austin, TX, USA, 11–17 July 2016; pp. 98–105. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://ccad.unc.edu.ar/ (accessed on 5 November 2022).
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard—7th Edition; CSLI Document M7–A7; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Cortez Marcolino, L.M.; Correia Pereira, A.H.; Guerra Pinto, J.; Mamone, L.A.; Ferreira Strixino, J. Cellular and metabolic changes after photodynamic therapy in leishmania promastigotes. Photodiagnosis Photodyn. Ther. 2021, 35, 102403. [Google Scholar] [CrossRef]
- Liu, J.; Sun, J.; Zhang, W.; Gao, K.; He, Z. HPLC determination of rifampicin and related compounds in pharmaceuticals using monolithic column. J. Pharm. Biomed. Anal. 2008, 46, 405–409. [Google Scholar] [CrossRef]
- ICH Harmonised Tripartite Guideline. Validation of Analytical Procedures: Text and Methodology. Q2 (R1), 2005.
- Jing, D.; Gu, Y.; Xia, H. Solid-State and Solution-Mediated Polymorphic Transformation of Rifampicin. Chem. Eng. Technol. 2018, 41, 1236–1243. [Google Scholar] [CrossRef]
- Popielec, A.; Loftsson, T. Effects of cyclodextrins on the chemical stability of drugs. Int. J. Pharm. 2017, 531, 532–542. [Google Scholar] [CrossRef]

















| Mode | VDW | ELE | PolSolv | NPolSolv | GAS | Total |
|---|---|---|---|---|---|---|
| mode-1 | −25.46 | 35.33 | 43.41 | −2.40 | −60.79 | −19.78 |
| mode-2 | −40.58 | −33.90 | 47.88 | −4.26 | −74.49 | 30.87 |
| mode-3 | −42.09 | −33.12 | 48.73 | −4.99 | −75.22 | −31.47 |
| mode-4 | −15.17 | −25.42 | 32.19 | −1.32 | −40.59 | −9.72 |
| mode-5 | −47.91 | −40.29 | 62.92 | −5.02 | −88.20 | −30.30 |
| mode-6 | −32.87 | −28.61 | 46.24 | −3.78 | −61.49 | −19.03 |
| Mode | VDW | ELE | PolSolv | NPolSolv | GAS | Total |
|---|---|---|---|---|---|---|
| mode-1 | −6.87 | −39.70 | 38.84 | −1.15 | −46.57 | −8.89 |
| mode-2 | −5.96 | −38.95 | 37.88 | −1.00 | −44.92 | −8.03 |
| Mode | VDW | ELE | PolSolv | NPolSolv | GAS | Total |
|---|---|---|---|---|---|---|
| mode-3.3.4 | −46.43 | −25.98 | 44.90 | −5.38 | −72.41 | −32.89 |
| VDW | ELE | PolSolv | NPolSolv | GAS | Total | |
|---|---|---|---|---|---|---|
| RIF:ARG | −11.10 | −38.02 | 42.45 | −1.58 | −49.13 | −8.26 |
| 1H-NMR Signals | |||
|---|---|---|---|
| RIF Protons | RIF:ARG | RIF:γ-CD | RIF:γ-CD:ARG |
| H33 | −0.0400 | −0.0470 | −0.0535 |
| H31 | −0.0265 | −0.0255 | −0.0645 |
| H32 | −0.0290 | −0.0310 | −0.0670 |
| H26 | −0.0240 | −0.0405 | −0.0670 |
| H24 | −0.0360 | −0.0455 | −0.0645 |
| H13 | −0.0300 | −0.0640 | −0.1330 |
| H22 | 0.0175 | −0.0060 | 0.0845 |
| H14 | −0.0290 | −0.0190 | −0.0430 |
| H30 | −0.0130 | −0.0440 | −0.0440 |
| H36 | −0.0310 | −0.0360 | −0.0630 |
| H20 | −0.0255 | −0.0245 | −0.0695 |
| H4’ | 0.0360 | 0.1280 | −0.3325 |
| H37 | −0.0320 | −0.0335 | −0.0550 |
| H3’–H5’ | −0.0015 | −0.0220 | −0.0685 |
| H2’–H6’ | −0.0750 | −0.0800 | −0.0315 |
| H27 | −0.0295 | −0.0335 | −0.0725 |
| H21 | −0.0310 | 0.0060 | −0.0260 |
| H28 | −0.0175 | 0.0055 | −0.0795 |
| H19 | 0.0000 | −0.0065 | −0.0845 |
| H29 | 0.0905 | −0.0265 | −0.0720 |
| H17 | 0.0205 | −0.0125 | −0.0715 |
| H18 | 0.0035 | 0.0650 | −0.0830 |
| H1′ | −0.0180 | 0.0170 | −0.4670 |
| γ-CD protons | |||
| H4 | - | 0.0020 | −0.0235 |
| H2 | - | −0.0015 | −0.0380 |
| H5–H6 | - | −0.0140 | −0.0760 |
| H3 | - | −0.0040 | −0.0360 |
| H1 | - | −0.0060 | −0.0470 |
| ARG protons | |||
| H4–H3 | 0.0345 | - | 0.0395 |
| H5 | 0.0015 | - | −0.0410 |
| H2 | −0.0250 | - | −0.0785 |
| RIF Dissolved (%) at Each Sampling Time | f2 Values | ||||
|---|---|---|---|---|---|
| 15 min | 30 min | 60 min | 120 min | ||
| Pure RIF | 14 ± 2% | 30 ± 2% | 46 ± 3% | 63 ± 1% | |
| RIF:ARG | 15 min | 30 min | 60 min | 120 min | f2 |
| PM | 45 ± 2% | 71 ± 3% | 91 ± 2% | 94.3 ± 0.4% | 23 |
| KN | 51 ± 4% | 70 ± 2% | 84 ± 3% | 94 ± 1% | 24 |
| FD | 63 ± 3% | 73 ± 1% | 79 ± 1% | 78 ± 1% | 24 |
| RIF:γ-CD | 15 min | 30 min | 60 min | 120 min | f2 |
| PM | 31 ± 1% | 52 ± 3% | 71 ± 2% | 80 ± 1% | 35 |
| KN | 68 ± 1% | 80 ± 1% | 80 ± 2% | 82 ± 3% | 22 |
| FD | 35 ± 2% | 50 ± 3% | 61 ± 2% | 74 ± 2% | 41 |
| RIF:γ-CD:ARG | 15 min | 30 min | 60 min | 120 min | f2 |
| PM | 43 ± 1% | 58 ± 3% | 68 ± 2% | 71 ± 3% | 34 |
| KN | 67 ± 2% | 67 ± 5% | 68 ± 2% | 68 ± 1% | 26 |
| FD | 42 ± 1% | 63 ± 2% | 83 ± 7% | 93 ± 3% | 26 |
| Parameter | Acceptance Criteria | Calculated Values |
|---|---|---|
| Linearity | r2 > 0.998 | Range = 50–150 µg/mL Area = 29.627 × C (µg/mL) − 42.365 r2 = 0.9996 |
| Accuracy | Recovery 98–102% | n = 9 RIF (µg/mL) = 50, 100, 150 Average recovery (%) = 100, 100, 99 |
| Repeatability | CV < 2% | n = 9 RIF (µg/mL) = 50, 100, 150 CV (%) = 0.01, 0.38, 0.88 |
| Intermediate precision | CV < 3% | n = 9 RIF (µg/mL) = 50, 100, 150 CV (%) = 0.01, 1.19, 0.33 |
| DL | DL << 0.05% | DL = 0.001% (0.002 µg/mL) |
| QL | QL ≤ 0.05% | QL = 0.004% (0.006 µg/mL) |
| Solutions | kc × 10−3 (h−1) | t90 (h) |
|---|---|---|
| RIF | 30 ± 1 | 3.5 ± 0.1 |
| RIF:ARG | 32.0 ± 0.4 | 3.28 ± 0.04 |
| RIF:γ-CD | 37.1 ± 0.4 | 2.88 ± 0.03 |
| RIF:γ-CD:ARG | 31.9 ± 0.1 | 3.29 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan Córdoba, A.V.; Aiassa, V.; Dimmer, J.A.; Barrionuevo, C.N.; Quevedo, M.A.; Longhi, M.R.; Zoppi, A. Development and Characterization of Pharmaceutical Systems Containing Rifampicin. Pharmaceutics 2023, 15, 198. https://doi.org/10.3390/pharmaceutics15010198
Dan Córdoba AV, Aiassa V, Dimmer JA, Barrionuevo CN, Quevedo MA, Longhi MR, Zoppi A. Development and Characterization of Pharmaceutical Systems Containing Rifampicin. Pharmaceutics. 2023; 15(1):198. https://doi.org/10.3390/pharmaceutics15010198
Chicago/Turabian StyleDan Córdoba, Antonella V., Virginia Aiassa, Jesica A. Dimmer, Camila N. Barrionuevo, Mario A. Quevedo, Marcela R. Longhi, and Ariana Zoppi. 2023. "Development and Characterization of Pharmaceutical Systems Containing Rifampicin" Pharmaceutics 15, no. 1: 198. https://doi.org/10.3390/pharmaceutics15010198