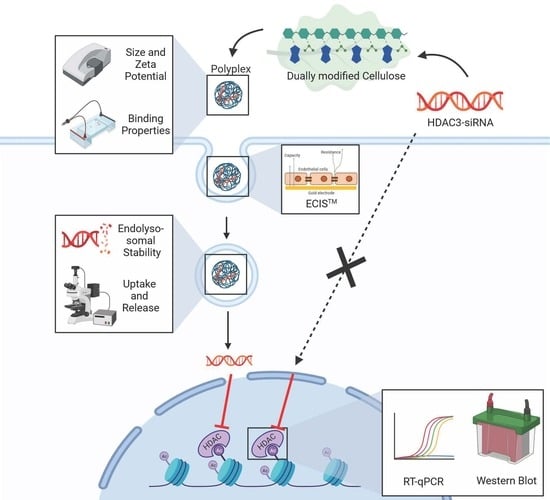
Dually Modified Cellulose as a Non-Viral Vector for the Delivery and Uptake of HDAC3 siRNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of 6-Deoxy-6-(2-aminoethyl) Amino Cellulose
- DSEDA: 0.54 (EDAC1); 0.81 (EDAC2, EDAC3).
- 1H NMR (250 MHz, D2O, δ): 7.93 (Harom), 7.61 (Harom), 5.29–3.09 (H1-6), 2.64 (H7,8).
- 13C NMR (63 MHz, D2O, δ): 129.67 (Carom), 125.69 (Carom), 102.67 (C1), 78.70–73.43 (C2, C3, C4, C5), 60.34 (C6), 49.12 (C6NH), 47.25 (C7), 39.30 (C8).
- EA: C 40.73; H 7.02; N 8.02; S 0.59; DSEDA = 0.54.
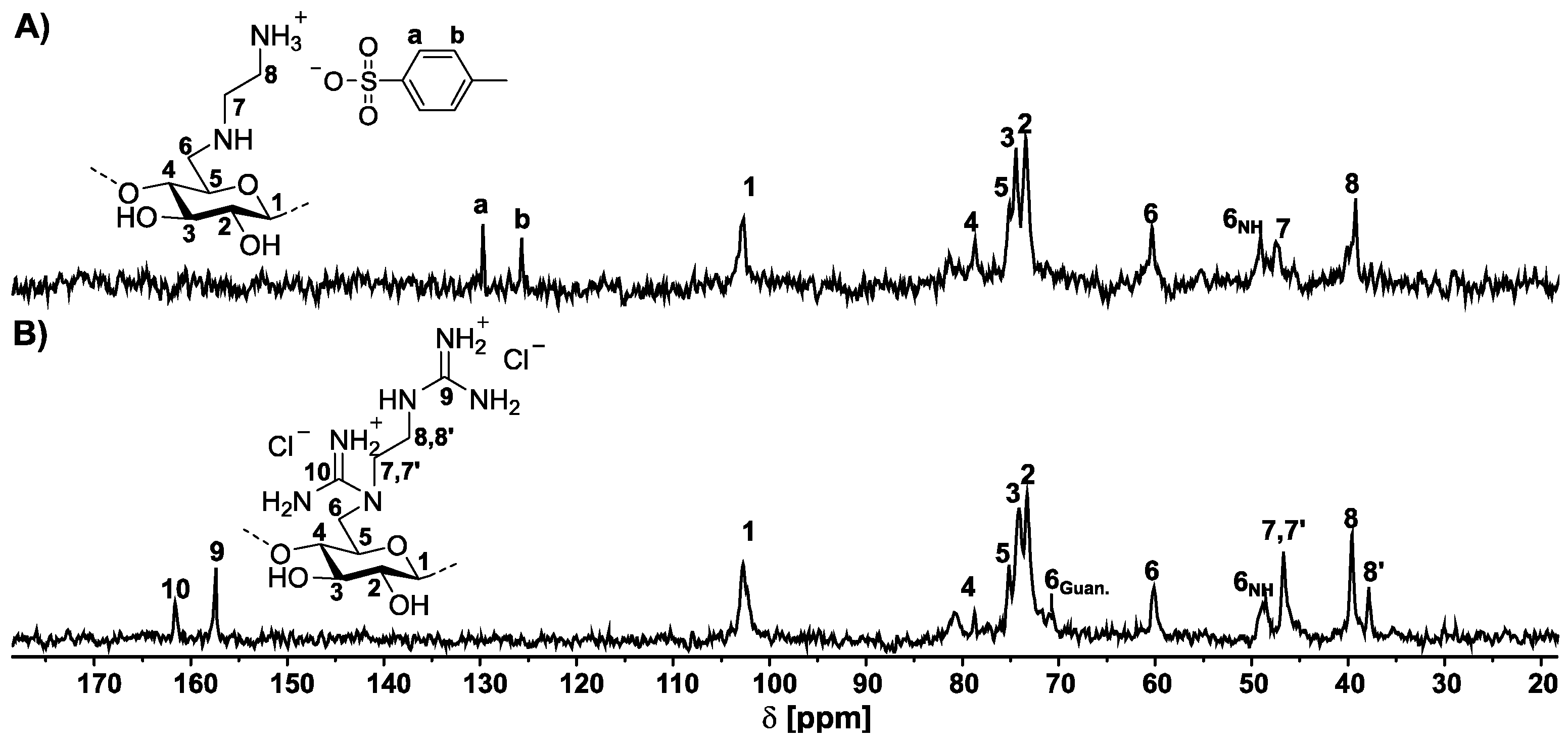
2.2. Synthesis of 6-Deoxy-6-(2-guanidiniumethyl) Amino Cellulose Chloride (Typical Example, GEDAC2)
- Yield: 0.64 g.
- 1H NMR (250 MHz, D2O, δ): 5.62–2.77 (H1-6; H7,8).
- 13C NMR (63 MHz, D2O, δ): 161.6 (C10), 157.4 (C9), 102.7 (C1), 83.8–65.7 (C2, C3, C4, C5), 70.9 (C6Guan.), 60.2 (C6OH), 48.6 (C6NH), 46.7 (C7), 39.7 (C8), 37.9 (C8′).
- EA: C 35.42%; H 6.48%; N 14.39%; Cl 9.95%.
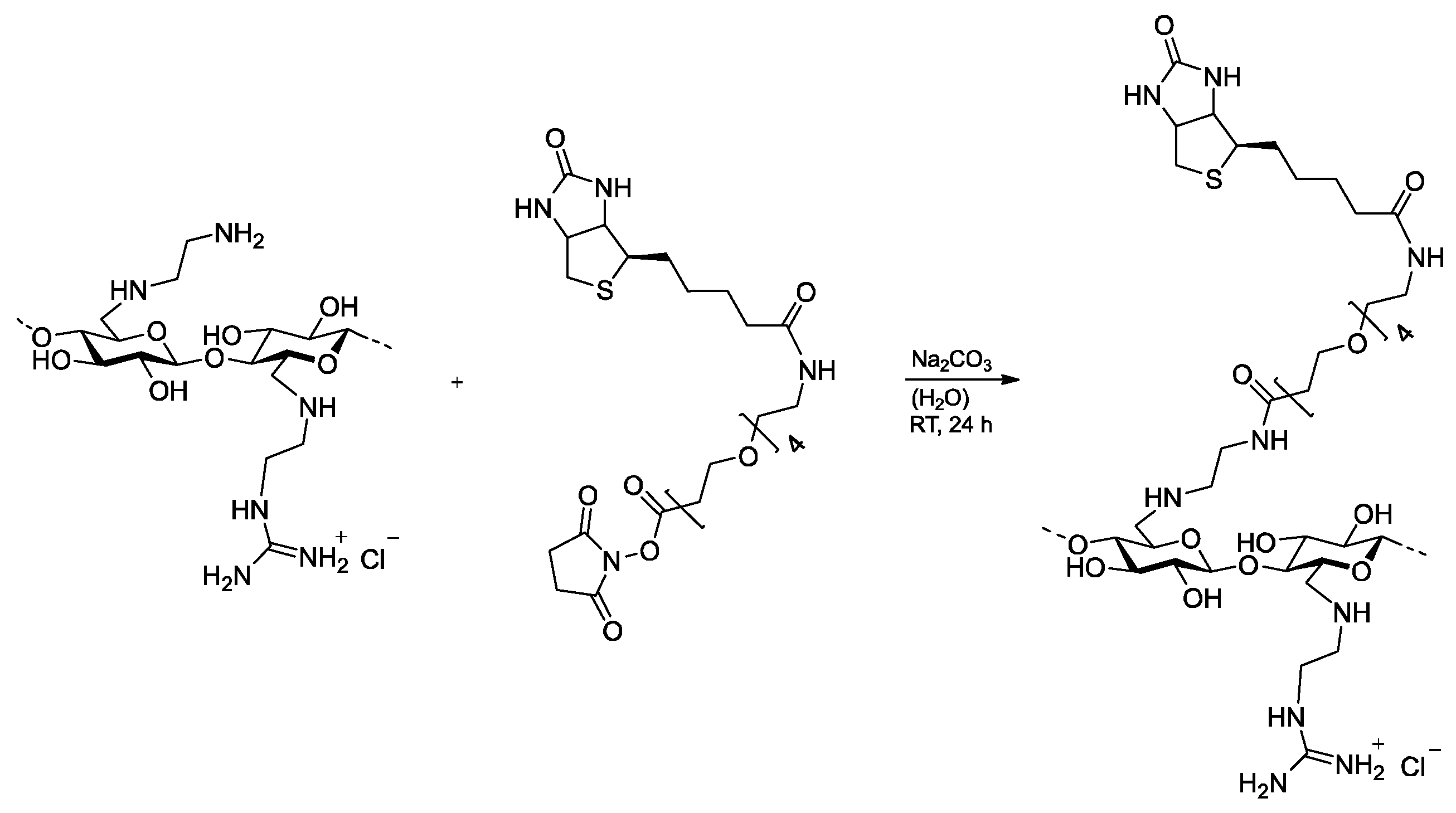
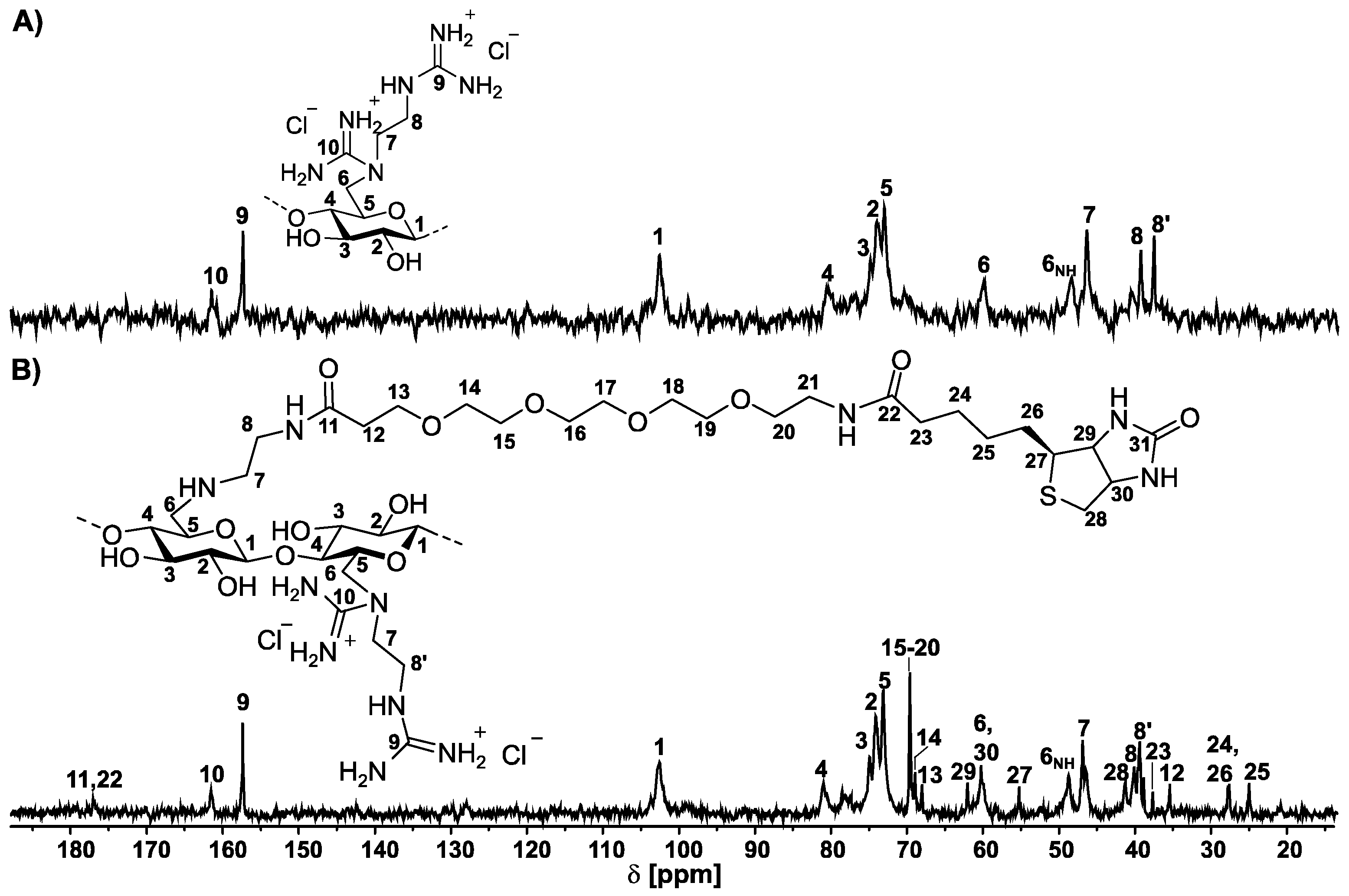
2.3. Biotinylation of 6-Deoxy-6-(2-aminoethyl) Amino Cellulose
- Yield: 0.25 g.
- 1H NMR (500 MHz, D2O, δ): 8.16 (NHBiotin), 7.83 (NHBiotin), 5.33–3.23 (H1-6, H7), 2.73 (H8), 2.53 (H23), 1.91 (H26), 1.68 (H25), 1.52 (H24).
- 13C NMR (101 MHz, D2O, δ): 177.1 (C11, C22), 161.6 (C10), 157.4 (C9), 102.7 (C1), 81.2 (C4) 75.2–73.4 (C2, C3, C5), 69.9 (C15–20), 69.2 (C14), 68.3 (C13), 62.4 (C29), 60.6 (C6, C30), 55.6 (C27), 49.0 (C6NH), 47.2 (C7), 46.8 (C7′), 41.6 (C28), 39.7 (C8), 38.1 (C8′), 35.8 (C23), 28.2 (C12), 28.0 (C24, C26), 25.4 (C25).
- EA: C 36.96%, H 6.50%, N 15.22%, S 0.61%.
- %N = percentage of nitrogen;
- MRU = molar mass of RU (162.14 g mol−1);
- MN = molar mass of nitrogen (14.00 g mol−1);
- nN sub = number of N in substituent;
- MEDA = molar mass of EDA (60.10 g mol−1);
- MH2O = molar mass of removed OH from repeating unit plus H from EDA (18.01 g mol−1).
- %S = percentage of sulfur;
- MRU = molar mass of RU (162.14 g mol−1);
- MEDA = molar mass of EDA (60.10 g mol−1);
- MH = molar mass of hydrogen (1.001 g mol−1);
- MS = molar mass of sulfur (32.06 g mol−1);
- MG = molar mass of the substituted part of guanidinium chloride (79.01 g mol−1);
- MBiotin-PEG4 = molar mass of Biotin-PEG4 (474.60 g mol−1);
2.4. Conductometric Titration
- DSEDA = DS of EDA;
- DSEDAn = new DS of EDA after guanylation.
- DSEDAn was calculated according to Equation (4):
- c = concentration of NaOH (0.01 M);
- MG = molar mass of the substituted part of guanidinium chloride (79.01 g mol−1);
- V = calculated titration volume of aqueous NaOH for non-guanylated amino moieties;
- MH = molar mass of hydrogen (1.001 g mol−1);
- MEDA = molar mass of EDA (60.10 g mol−1);
- MOH = molar mass of OH (17.00 g mol−1);
- MRU = molar mass of RU (162.14 g mol−1);
- mp = sample mass.
2.5. Size-Exclusion Chromatography (SEC) Measurement
2.6. Polyplex Formation
2.7. Binding Capacity
2.8. Particle Size and Zeta Potential Measurements
2.9. Endolysosomal Stability of Polyplexes
2.10. In Vitro Cell Culture
2.11. Cell Viability Assays
2.12. Barrier Integrity Measurement Based on Electric Cell-Substrate Impedance Sensing (ECISTM)
2.13. siRNA Transfection and Real-Time Quantitative PCR
- HDAC3 forward, 5′-GAGTTCTGCTCGCGTTACACAG-3′-;
- HDAC3 reverse, 5′-CGTTGACATAGCAGAAGCCAGAG-3′;
- Rps2 forward, 5′-GCACCAGGTTCAAGGCATTTG-3′;
- Rps2 reverse, 5′-TCTTGTTCCCCCAGTAGCCT-3′.
2.14. Western Blot
2.15. Immunofluorescence (IF)
2.16. Statistical Analyses
3. Results and Discussion
3.1. Guanylation of 6-Deoxy-6-(2-aminoethyl) Amino Cellulose
3.2. NMR Spectroscopic Characterization of 6-Deoxy-6-(2-guanidiniumethyl) Amino Cellulose Chloride
3.3. Biotinylation of 6-Deoxy-6-(2-guanidiniumethyl) Amino Cellulose
3.4. Binding Capacity
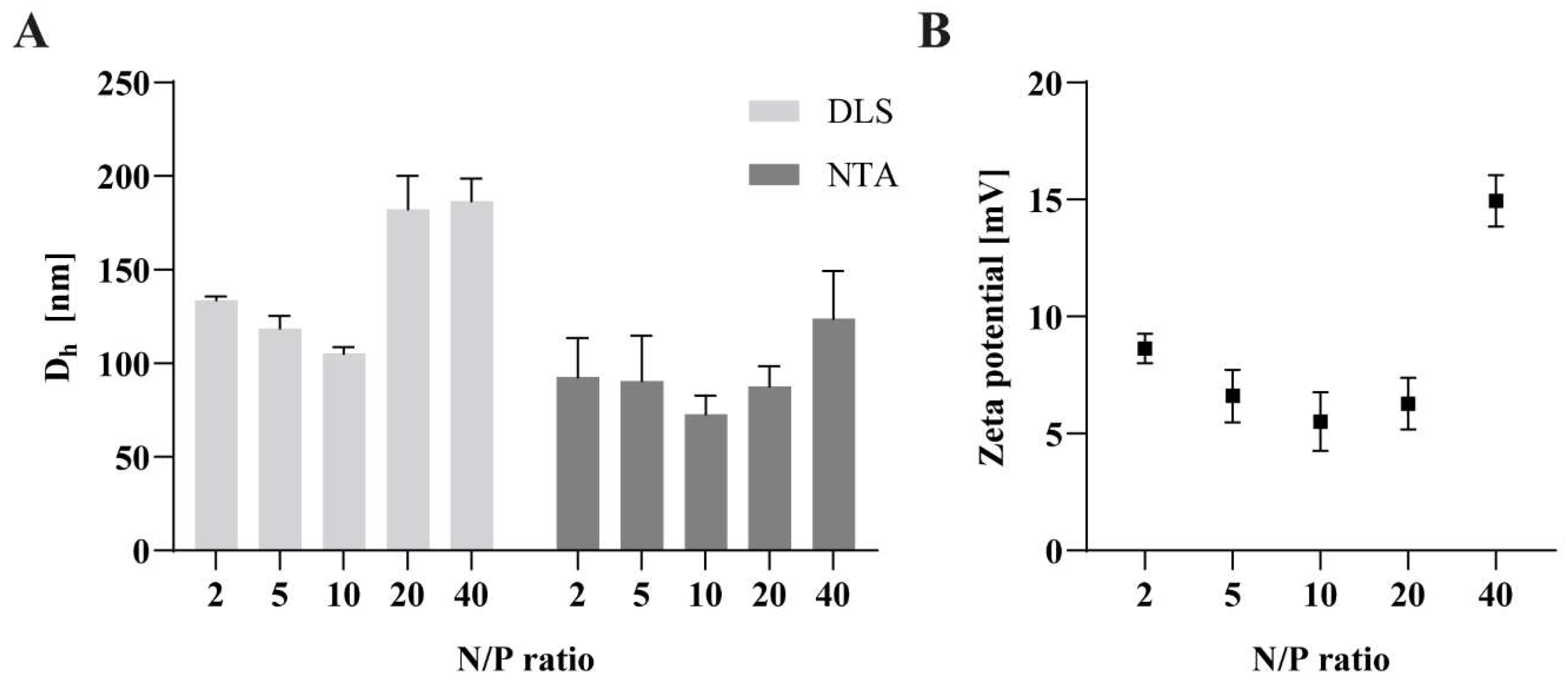
3.5. Particle Size and Zeta Potential
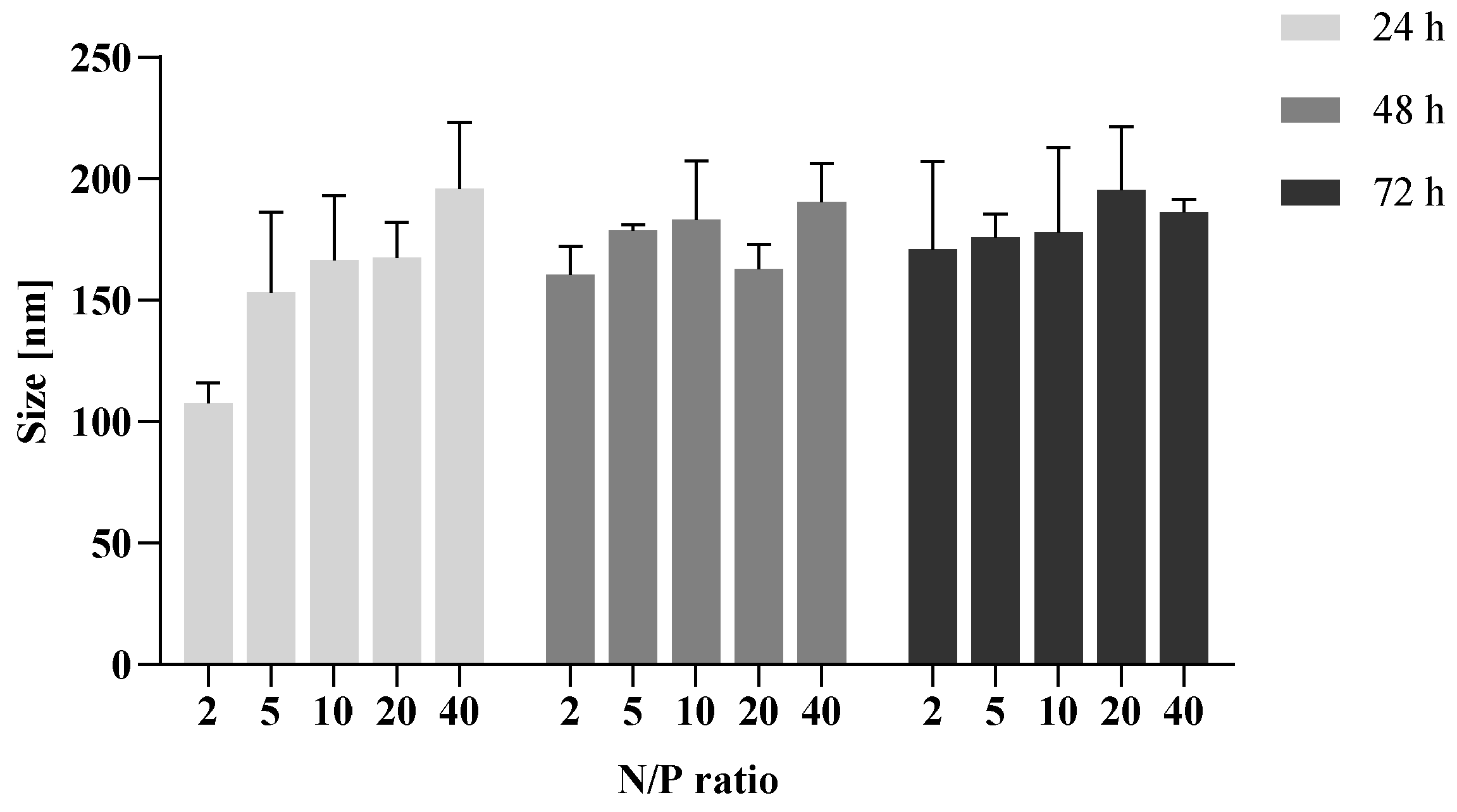
3.6. Endolysosomal Stability
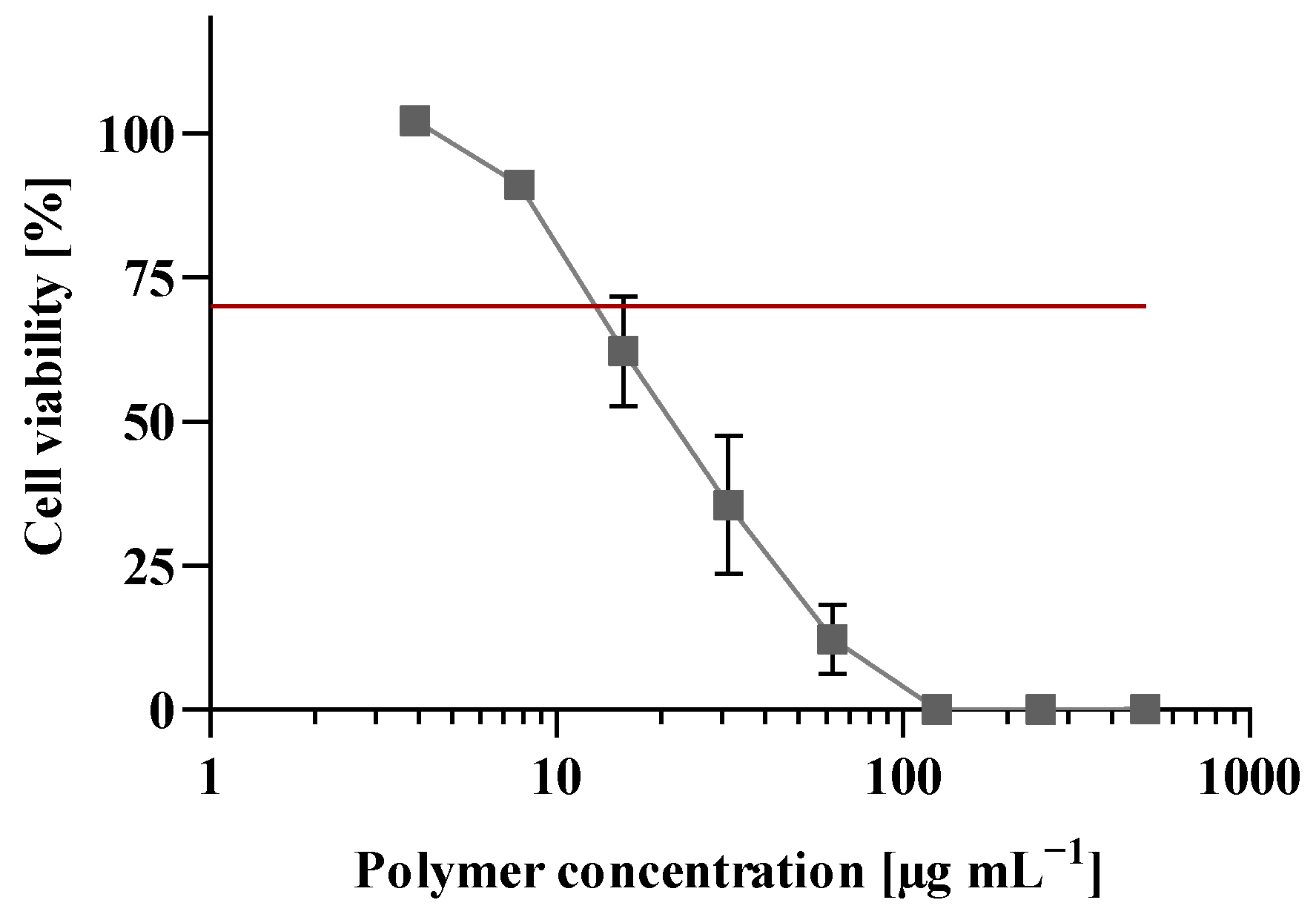
3.7. Cell Viability Assay
3.8. Effect of Polyplexes on Endothelial Barrier Integrity
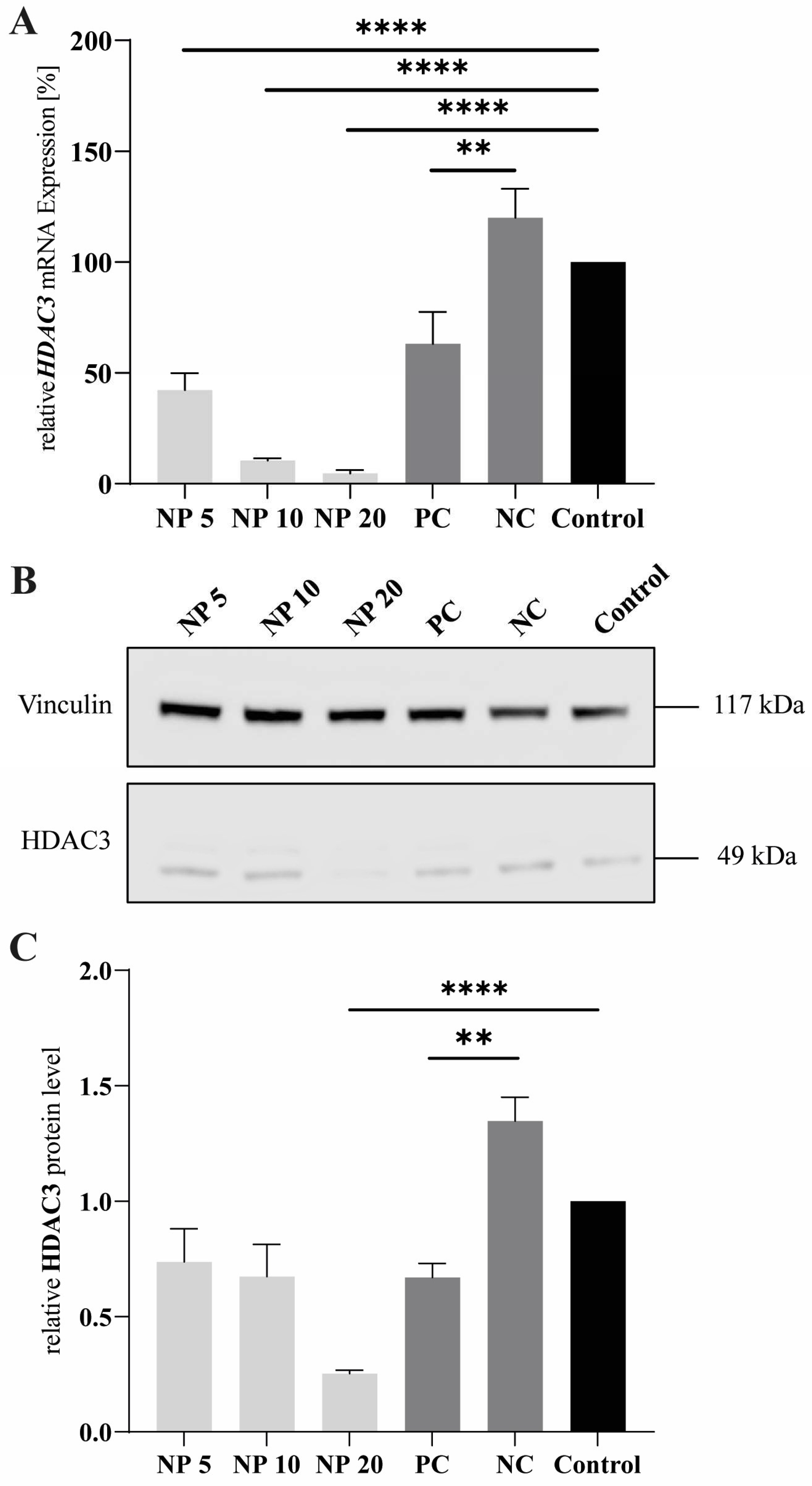
3.9. Polyplex Transfection
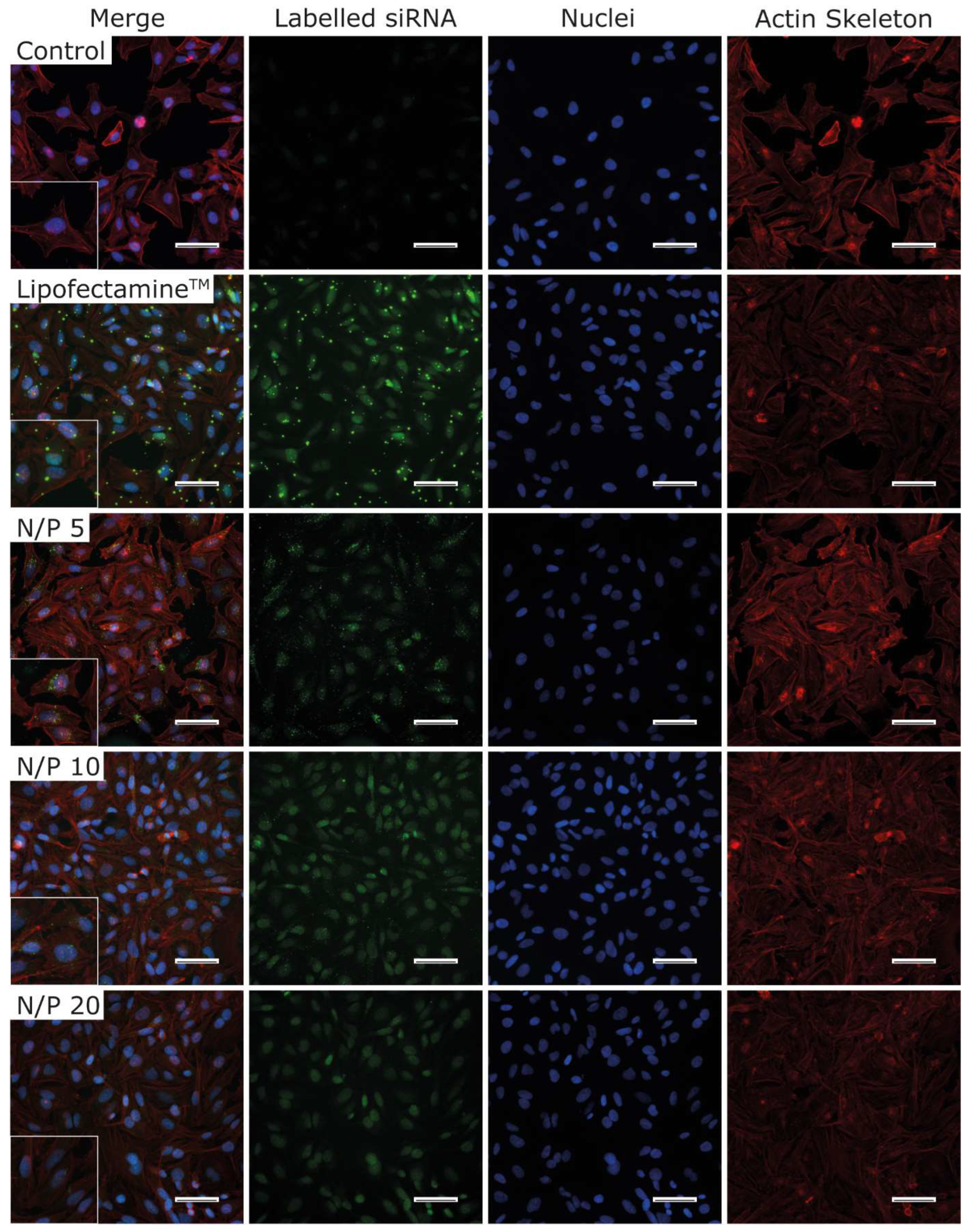
3.10. Immunofluorescence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.J.; Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.D. RNA Interference and Cancer Therapy; Springer Science + Business Media: New York, NY, USA, 2019. [Google Scholar] [CrossRef]
- Layzer, J.M.; McCaffrey, A.P.; Tanner, A.K.; Huang, Z.; Kay, M.A.; Sullenger, B.A. In vivo activity of nuclease-resistant siRNAs. RNA 2004, 10, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.M.; Dennahy, I.S.; Bhatti, U.F.; Biesterveld, B.E.; Graham, N.J.; Li, Y.; Alam, H.B. Histone Deacetylase Inhibitors: A Novel Strategy in Trauma and Sepsis. Shock 2019, 52, 300–306. [Google Scholar] [CrossRef]
- Roger, T.; Lugrin, J.; Le Roy, D.; Goy, G.; Mombelli, M.; Koessler, T.; Ding, X.C.; Chanson, A.L.; Reymond, M.K.; Miconnet, I.; et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 2011, 117, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Chen, X.; Barozzi, I.; Termanini, A.; Prosperini, E.; Recchiuti, A.; Dalli, J.; Mietton, F.; Matteoli, G.; Hiebert, S.; Natoli, G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. USA 2012, 109, E2865–E2874. [Google Scholar] [CrossRef]
- Chen, S.; Bellew, C.; Yao, X.; Stefkova, J.; Dipp, S.; Saifudeen, Z.; Bachvarov, D.; El-Dahr, S.S. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J. Biol. Chem. 2011, 286, 32775–32789. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, W. Histone deacetylase 3 (HDAC3) as an important epigenetic regulator of kidney diseases. J. Mol. Med. 2022, 100, 43–51. [Google Scholar] [CrossRef]
- Aggarwal, R.; Targhotra, M.; Kumar, B.; Sahoo, P.K.; Chauhan, M.K. Chauhan Polyplex: A Promising Gene Delivery System. Int. J. Pharm. Sci. Nanotechnol. 2019, 12, 4681–4686. [Google Scholar]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Hack, F.J.; Cokca, C.; Stadter, S.; Hülsmann, J.; Peneva, K.; Fischer, D. Indole, Phenyl, and Phenol Groups: The Role of the monomer on Gene Delivery in Guanidinium Containing Methacrylamide Terpolymers. Macromol. Rapid Commun. 2020, 42, e2000580. [Google Scholar] [CrossRef] [PubMed]
- Zink, M.; Hotzel, K.; Schubert, U.S.; Heinze, T.; Fischer, D. Amino Acid-Substituted Dextran-Based Non-Viral Vectors for Gene Delivery. Macromol. Biosci. 2019, 19, e1900085. [Google Scholar] [CrossRef]
- Rowe, R.C.; Sheskey, P.J.; Quinn, M.E. Handbook of Pharmaceutical Exipients, 6th ed.; Pharmaceutical Press: London, UK, 2009. [Google Scholar]
- Xu, F.; Ping, Y.; Ma, J.; Tang, G.; Yang, W.; Li, J.; Kang, E.; Neoh, K. Comb-shaped copolymers composed of hydroxypropyl cellulose backbones and cationic poly ((2-dimethyl amino) ethyl methacrylate) side chains for gene delivery. Bioconjugate Chem. 2009, 20, 1449–1458. [Google Scholar] [CrossRef]
- Stanzl, E.G.; Trantow, B.M.; Vargas, J.R.; Wender, P.A. Fifteen Years of Cell-Penetrating, Guanidinium-Rich Molecular Transporters: Basic Science, Research Tools, and Clinical Applications. Acc. Chem. Res. 2013, 46, 2944–2954. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski-Daspit, A.S.; Kauffman, A.C.; Bracaglia, L.G.; Saltzman, W.M. Polymeric vehicles for nucleic acid delivery. Adv. Drug. Deliv. Rev. 2020, 156, 119–132. [Google Scholar] [CrossRef]
- Suzuki, T.; Futaki, S.; Niwa, M.; Tanaka, S.; Ueda, K.; Sugiura, Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J. Biol. Chem. 2002, 277, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Rothbard, J.B.; Jessop, T.C.; Lewis, R.S.; Murray, B.A.; Wender, P.A. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004, 126, 9506–9507. [Google Scholar] [CrossRef]
- Tabujew, I.; Freidel, C.; Krieg, B.; Helm, M.; Koynov, K.; Mullen, K.; Peneva, K. The guanidinium group as a key part of water-soluble polymer carriers for siRNA complexation and protection against degradation. Macromol. Rapid Commun. 2014, 35, 1191–1197. [Google Scholar] [CrossRef]
- Wender, P.A.; Galliher, W.C.; Goun, E.A.; Jones, L.R.; Pillow, T.H. The design of guanidinium-rich transporters and their internalization mechanisms. Adv. Drug. Deliv. Rev. 2008, 60, 452–472. [Google Scholar] [CrossRef]
- Tahir, S.; Badshah, A.; Hussain, R.A. Guanidines from ‘toxic substances’ to compounds with multiple biological applications–Detailed outlook on synthetic procedures employed for the synthesis of guanidines. Bioorg. Chem. 2015, 59, 39–79. [Google Scholar] [CrossRef]
- Eswaran, L.; Kazimirsky, G.; Byk, G. New Biocompatible Nanohydrogels of Predefined Sizes for Complexing Nucleic Acids. Pharmaceutics 2023, 15, 332. [Google Scholar] [CrossRef]
- Eswaran, L.; Kazimirsky, G.; Yehuda, R.; Byk, G. A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes. Pharmaceutics 2023, 15, 961. [Google Scholar] [CrossRef]
- Bernatowicz, M.S.; Wu, Y.; Matsueda, G.R. 1H-Pyrazole-1-carboxamidine hydrochloride an attractive reagent for guanylation of amines and its application to peptide synthesis. J. Org. Chem. 1992, 57, 2497–2502. [Google Scholar] [CrossRef]
- Wagman, A.S.; Boyce, R.S.; Brown, S.P.; Fang, E.; Goff, D.; Jansen, J.M.; Le, V.P.; Levine, B.H.; Ng, S.C.; Ni, Z.J.; et al. Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3. J. Med. Chem. 2017, 60, 8482–8514. [Google Scholar] [CrossRef]
- Heinze, T.; Pfeifer, A.; Koschella, A.; Schaller, J.; Meister, F. Solvent-free synthesis of 6-deoxy-6-(ω-aminoalkyl)amino cellulose. J. Appl. Polym. Sci. 2016, 133, 43987. [Google Scholar] [CrossRef]
- Chen, G.; Wang, K.; Wang, Y.; Wu, P.; Sun, M.; Oupicky, D. Fluorination Enhances Serum Stability of Bioreducible Poly(amido amine) Polyplexes and Enables Efficient Intravenous siRNA Delivery. Adv. Healthc. Mater. 2018, 7, 1700978. [Google Scholar] [CrossRef]
- Cokca, C.; Hack, F.J.; Costabel, D.; Herwig, K.; Hulsmann, J.; Then, P.; Heintzmann, R.; Fischer, D.; Peneva, K. PEGylation of Guanidinium and Indole Bearing Poly(methacrylamide)s—Biocompatible Terpolymers for pDNA Delivery. Macromol. Biosci. 2021, 21, e2100146. [Google Scholar] [CrossRef] [PubMed]
- Fingernagel, J.; Boye, S.; Kietz, A.; Hobel, S.; Wozniak, K.; Moreno, S.; Janke, A.; Lederer, A.; Aigner, A.; Temme, A.; et al. Mono- and Polyassociation Processes of Pentavalent Biotinylated PEI Glycopolymers for the Fabrication of Biohybrid Structures with Targeting Properties. Biomacromolecules 2019, 20, 3408–3424. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.; Hansen, S.; Heydenreich, A.V.; Kristensen, H.G.; Poulsen, H.S. Solid lipid nanoparticles can effectively bind DNA, streptavidin and biotinylated ligands. Eur. J. Pharm. Biopharm. 2006, 62, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Jugel, W.; Aigner, A.; Michen, S.; Hagstotz, A.; Ewe, A.; Appelhans, D.; Schackert, G.; Temme, A.; Tietze, S. Targeted RNAi of BIRC5/Survivin Using Antibody-Conjugated Poly(Propylene Imine)-Based Polyplexes Inhibits Growth of PSCA-Positive Tumors. Pharmaceutics 2021, 13, 676. [Google Scholar] [CrossRef] [PubMed]
- Haladjova, E.; Smolíček, M.; Ugrinova, I.; Momekova, D.; Shestakova, P.; Kroneková, Z.; Kronek, J.; Rangelov, S. DNA delivery systems based on copolymers of poly (2-methyl-2-oxazoline) and polyethyleneimine: Effect of polyoxazoline moieties on the endo-lysosomal escape. J. Appl. Polym. Sci. 2020, 137, 49400. [Google Scholar] [CrossRef]
- Dennhardt, S.; Finke, K.R.; Huwiler, A.; Coldewey, S.M. Sphingosine-1-phosphate promotes barrier-stabilizing effects in human microvascular endothelial cells via AMPK-dependent mechanisms. Biochim. Biophys. Acta. Mol. Basis. Dis. 2019, 1865, 774–781. [Google Scholar] [CrossRef] [PubMed]
- DIN EN ISO 10993-5:2009-10; Biological Evaluation of Medical Devices—Part 5: Test for In Vitro Cytotoxicity. European Standard EN ISO 10993–5. ISO: Brussels, Belgium, 2009.
- Howard Bradbury, J.; Johnson, R.N. Assignment of the 13C NMR spectrum of biotin. J. Magn. Reson. 1979, 35, 217–222. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. Behr A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Wang, X.L.; Zhang, P.C.; Liu, Z.L.; Zhuo, R.X.; Mao, H.Q.; Leong, K.W. Galactosylated ternary DNA/polyphosphoramidate nanoparticles mediate high gene transfection efficiency in hepatocytes. J. Control. Release 2005, 102, 749–763. [Google Scholar] [CrossRef]
- Layek, B.; Singh, J. N-hexanoyl, N-octanoyl and N-decanoyl chitosans: Binding affinity, cell uptake, and transfection. Carbohydr. Polym. 2012, 89, 403–410. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA 2003, 9, 1034–1048. [Google Scholar] [CrossRef]
- Malek, A.; Czubayko, F.; Aigner, A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. J. Drug Target. 2008, 16, 124–139. [Google Scholar] [CrossRef]
- Zhu, D.; Yan, H.; Zhou, Z.; Tang, J.; Liu, X.; Hartmann, R.; Parak, W.J.; Feliu, N.; Shen, Y. Detailed investigation on how the protein corona modulates the physicochemical properties and gene delivery of polyethylenimine (PEI) polyplexes. Biomater. Sci. 2018, 6, 1800–1817. [Google Scholar] [CrossRef]
- Maguire, C.M.; Rosslein, M.; Wick, P.; Prina-Mello, A. Characterisation of particles in solution—A perspective on light scattering and comparative technologies. Sci. Technol. Adv. Mater. 2018, 19, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Win, K.Y.; Feng, S.S. Effects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugs. Biomaterials 2005, 26, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Foged, C.; Brodin, B.; Frokjaer, S.; Sundblad, A. Particle size and surface charge affect particle uptake by human dendritic cells in an in vitro model. Int. J. Pharm. 2005, 298, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; May, S. Release of cationic polymer-DNA complexes from the endosome: A theoretical investigation of the proton sponge hypothesis. J. Chem. Phys. 2008, 129, 185105. [Google Scholar] [CrossRef] [PubMed]
- Baskin, D.S.; Ngo, H.; Didenko, V.V. Thimerosal Induces DNA Breaks, Caspase-3 Activation, Membrane Damage, and Cell Death in Cultured Human Neurons and Fibroblasts. J. Toxicol. Sci. 2003, 74, 361–368. [Google Scholar] [CrossRef]
- Fischer, D.; Dusek, N.; Hotzel, K.; Heinze, T. The Role of Formamidine Groups in Dextran Based Nonviral Vectors for Gene Delivery on Their Physicochemical and Biological Characteristics. Macromol. Biosci. 2020, 21, e2000220. [Google Scholar] [CrossRef]
- Lan, B.; Wu, J.; Li, N.; Pan, C.; Yan, L.; Yang, C.; Zhang, L.; Yang, L.; Ren, M. Hyperbranched cationic polysaccharide derivatives for efficient siRNA delivery and diabetic wound healing enhancement. Int. J. Biol. Macromol. 2020, 154, 855–865. [Google Scholar] [CrossRef]
- Bozzuto, G.; D’Avenio, G.; Condello, M.; Sennato, S.; Battaglione, E.; Familiari, G.; Molinari, A.; Grigioni, M. Label-free cell based impedance measurements of ZnO nanoparticles-human lung cell interaction: A comparison with MTT, NR, Trypan blue and cloning efficiency assays. J. Nanobiotechnol. 2021, 19, 306. [Google Scholar] [CrossRef]
- Nahid, M.A.; Campbell, C.E.; Fong KS, K.; Barnhill, J.C.; Washington, M.A. An evaluation of the impact of clinical bacterial isolates on epithelial cell monolayer integrity by the electric Cell-Substrate Impedance Sensing (ECIS) method. J. Microbiol. Methods 2020, 169, 105833. [Google Scholar] [CrossRef]
- Park, I.; Nguyen, T.; Park, J.; Yoo, A.Y.; Park, J.K.; Cho, S. Impedance Characterization of Chitosan Cytotoxicity to MCF-7 Breast Cancer Cells Using a Multidisc Indium Tin Oxide Microelectrode Array. J. Electrochem. Soc. 2018, 165, B55–B59. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, S.; Zhang, J.; Wang, H.; Zou, Q.; Wu, L. Stealthy nanoparticles protect endothelial barrier from leakiness by resisting the absorption of VE-cadherin. Nanoscale 2021, 13, 12577–12586. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Setyawati, M.I.; Tee, J.K.; Ding, X.; Wang, J.; Nga, M.E.; Ho, H.K.; Leong, D.T. Nanoparticles promote in vivo breast cancer cell intravasation and extravasation by inducing endothelial leakiness. Nat. Nanotechnol. 2019, 14, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for delivery of nucleic acids. Adv. Drug Deliv. Rev. 2013, 65, 1234–1270. [Google Scholar] [CrossRef] [PubMed]
- Pretzer, I.; Bushiri, D.; Weberskirch, R. Biotin-Functionalized Block Catiomers as an Active Targeting Approach in Gene Delivery. Macromol. Mater. Eng. 2022, 308, 2200627. [Google Scholar] [CrossRef]
- Smith, S.A.; Selby, L.I.; Johnston AP, R.; Such, G.K. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem. 2018, 30, 263–272. [Google Scholar] [CrossRef]
- Bus, T.; Traeger, A.; Schubert, U.S. The great escape: How cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B 2018, 6, 6904–6918. [Google Scholar] [CrossRef]
- Yang, W.M.; Tsai, S.C.; Wen, Y.D.; Fejer, G.; Seto, E. Functional domains of histone deacetylase-3. J. Biol. Chem. 2002, 277, 9447–9454. [Google Scholar] [CrossRef]
- Berezhna, S.Y.; Supekova, L.; Supek, F.; Schultz, P.G.; Deniz, A.A. siRNA in human cells selectively localizes to target RNA sites. Proc. Natl. Acad. Sci. USA 2006, 103, 7682–7687. [Google Scholar] [CrossRef]
- Gagnon, K.T.; Li, L.; Chu, Y.; Janowski, B.A.; Corey, D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014, 6, 211–221. [Google Scholar] [CrossRef]
- Bus, T.; Englert, C.; Reifarth, M.; Borchers, P.; Hartlieb, M.; Vollrath, A.; Hoeppener, S.; Traeger, A.; Schubert, U.S. 3rd generation poly(ethylene imine)s for gene delivery. J. Mater. Chem. B 2017, 5, 1258–1274. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhou, L.; Yue, Q.; Liu, Q.; Cai, X.; Xiao, W.; Hai, L.; Guo, L.; Wu, Y. Liposomes modified with double-branched biotin: A novel and effective way to promote breast cancer targeting. Bioorg. Med. Chem. 2019, 27, 3115–3127. [Google Scholar] [CrossRef] [PubMed]

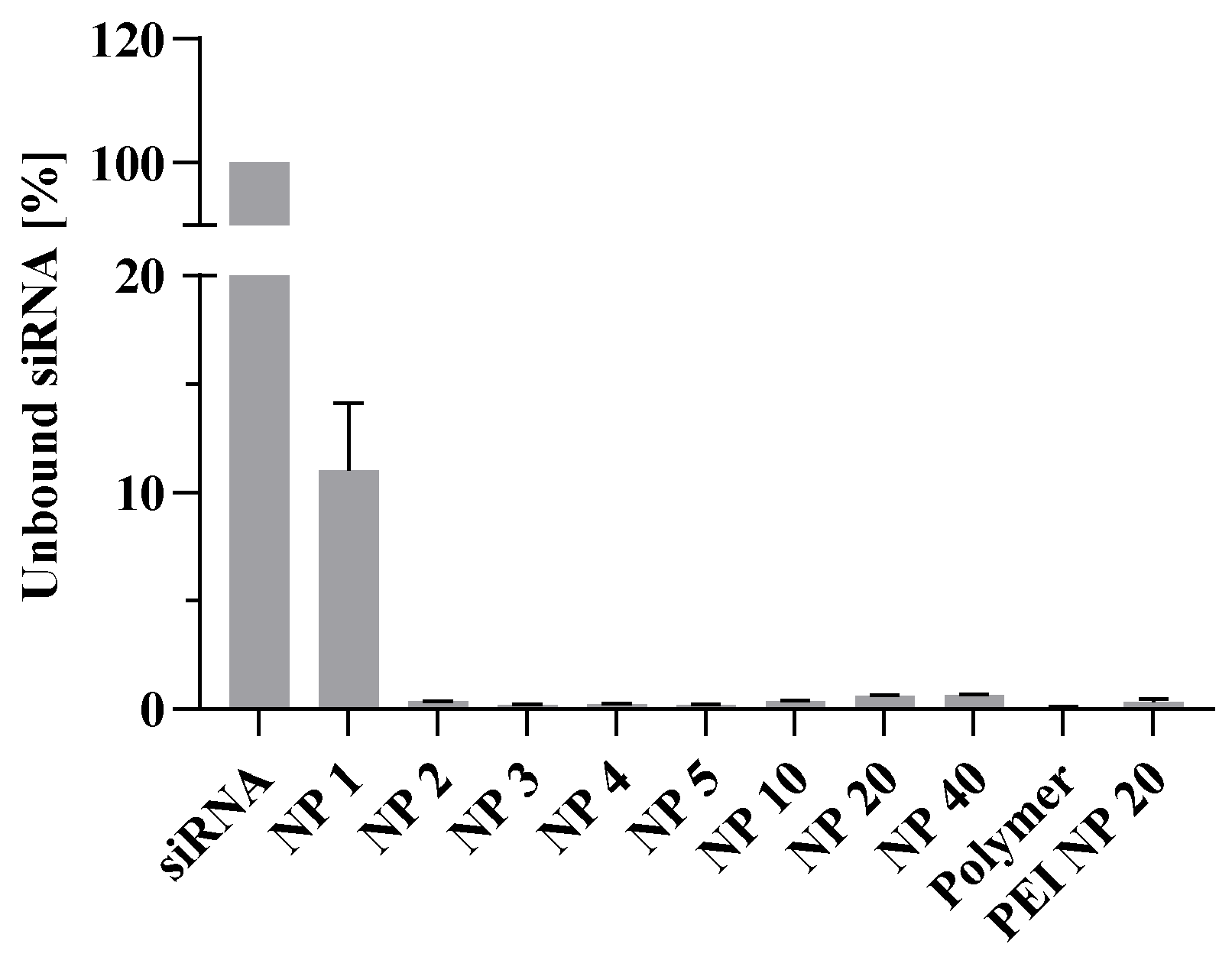

| Conditions | Sample | Results | ||
|---|---|---|---|---|
| DSEDA | T [°C] | Degree of Guanylation DG (a) | Conversion of EDA [%] | |
| 0.54 | 60 | GEDAC1 | 0.25 | 46.3 |
| 0.54 | 80 | GEDAC2 | 0.33 | 61.1 |
| 0.81 | 80 | GEDAC3 | 0.62 | 76.5 |
| 0.81 | 80 | GEDAC4 | 0.69 | 85.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hülsmann, J.; Lindemann, H.; Wegener, J.; Kühne, M.; Godmann, M.; Koschella, A.; Coldewey, S.M.; Heinze, T.; Heinzel, T. Dually Modified Cellulose as a Non-Viral Vector for the Delivery and Uptake of HDAC3 siRNA. Pharmaceutics 2023, 15, 2659. https://doi.org/10.3390/pharmaceutics15122659
Hülsmann J, Lindemann H, Wegener J, Kühne M, Godmann M, Koschella A, Coldewey SM, Heinze T, Heinzel T. Dually Modified Cellulose as a Non-Viral Vector for the Delivery and Uptake of HDAC3 siRNA. Pharmaceutics. 2023; 15(12):2659. https://doi.org/10.3390/pharmaceutics15122659
Chicago/Turabian StyleHülsmann, Juliana, Henry Lindemann, Jamila Wegener, Marie Kühne, Maren Godmann, Andreas Koschella, Sina M. Coldewey, Thomas Heinze, and Thorsten Heinzel. 2023. "Dually Modified Cellulose as a Non-Viral Vector for the Delivery and Uptake of HDAC3 siRNA" Pharmaceutics 15, no. 12: 2659. https://doi.org/10.3390/pharmaceutics15122659