Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Physical Measurements
2.2. Synthesis
2.2.1. Synthesis of the Ligands
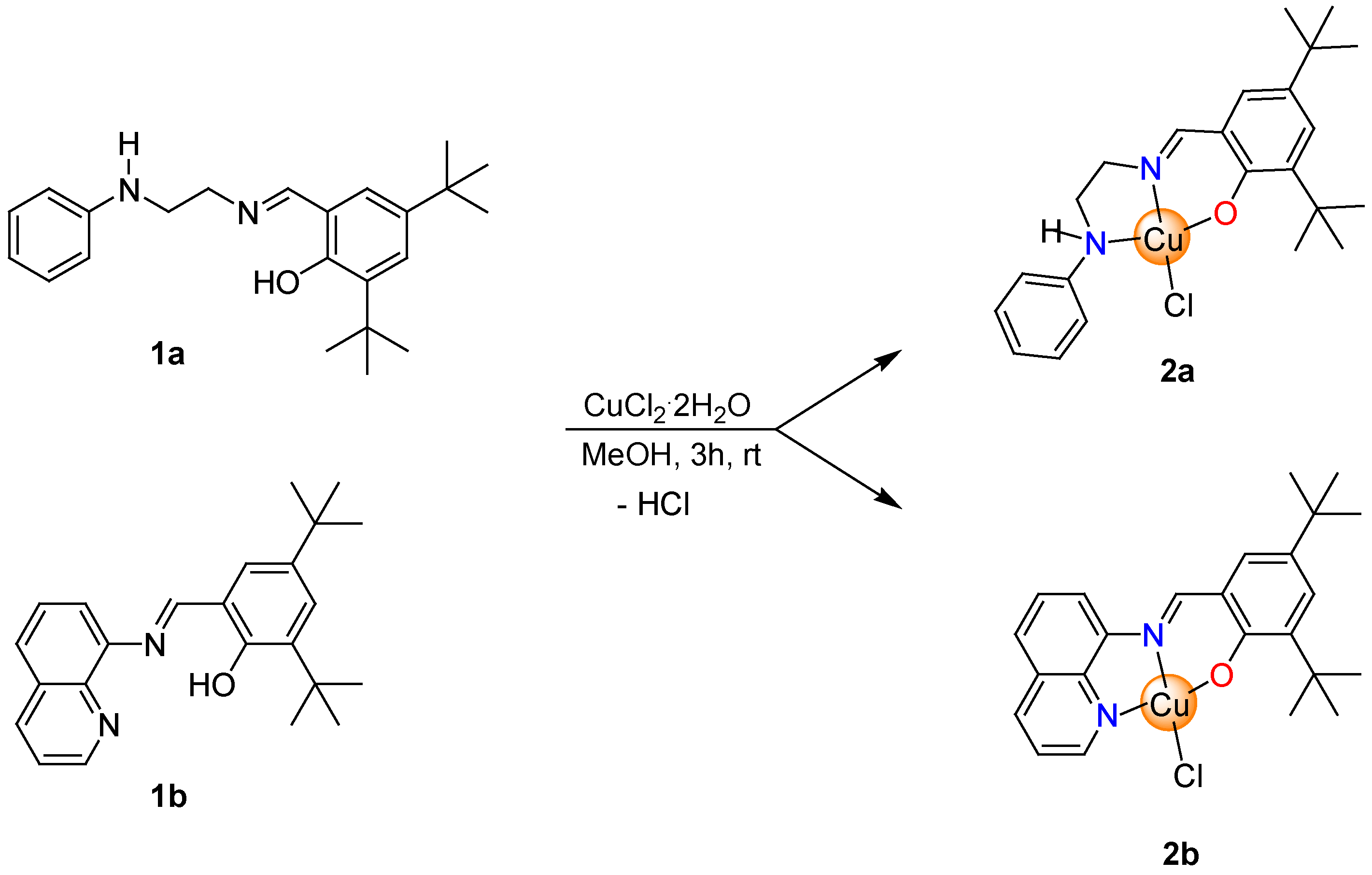
2.2.2. Synthesis of Copper Complexes
2.3. Single Crystal X-ray Diffraction
2.4. Theoretical Methods
2.5. Molecular Docking
2.6. Studies on the Stability of the Complexes in Solution
2.7. Biological Activities Studies
2.7.1. Determination of Free Radical Scavenging Capacity
2.7.2. Cell Culture and Treatments
2.7.3. Cell Viability Assessment by MTT Assay
2.7.4. Clonogenic Assay
2.7.5. Alkaline Comet Assay
2.8. Statistical Analysis
3. Results and Discussion
3.1. Chemical Section
3.1.1. Synthesis and Characterization of the Compounds
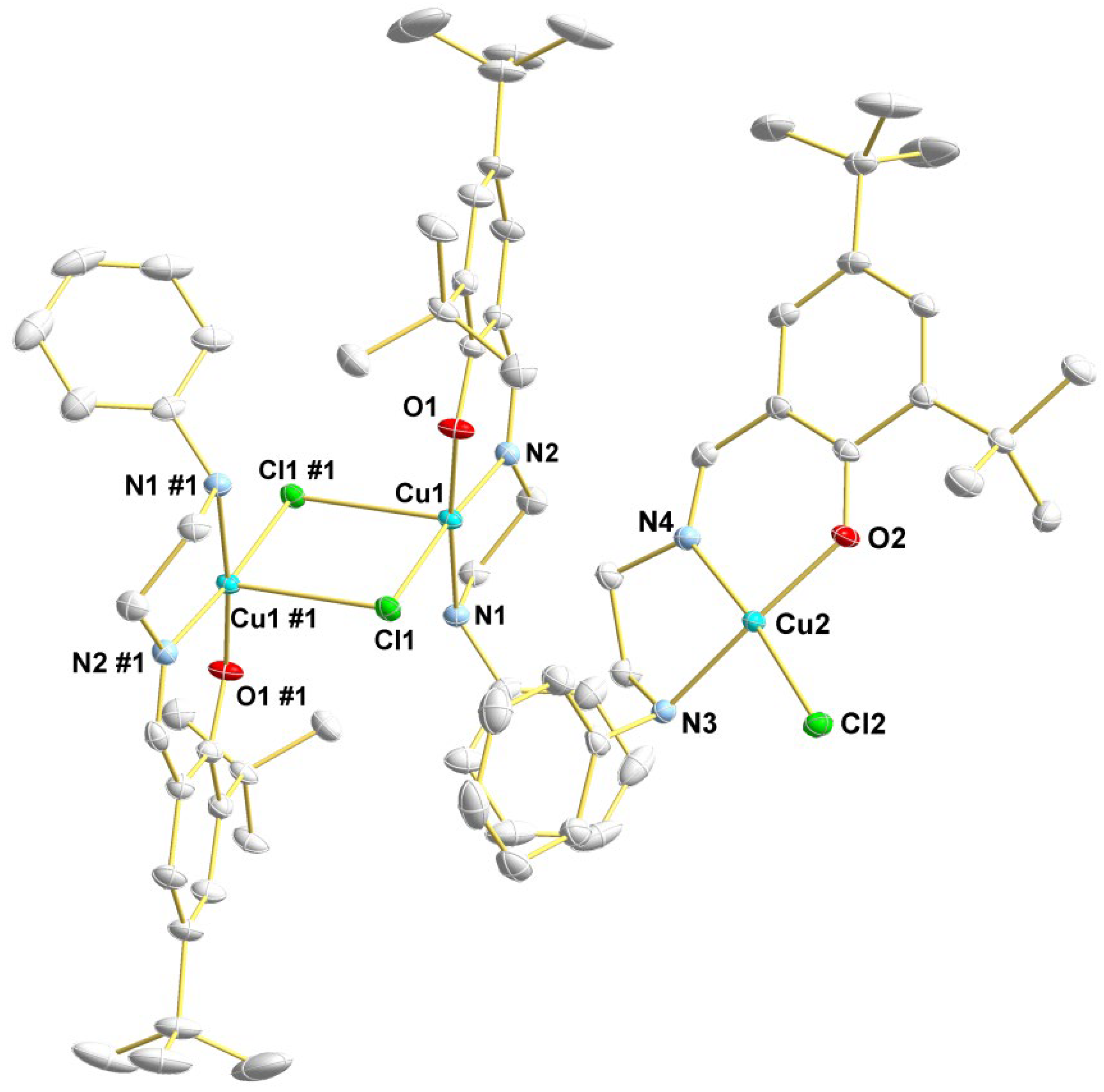
3.1.2. Description of the Crystal Structure of Complex 2a
3.1.3. DFT Calculations
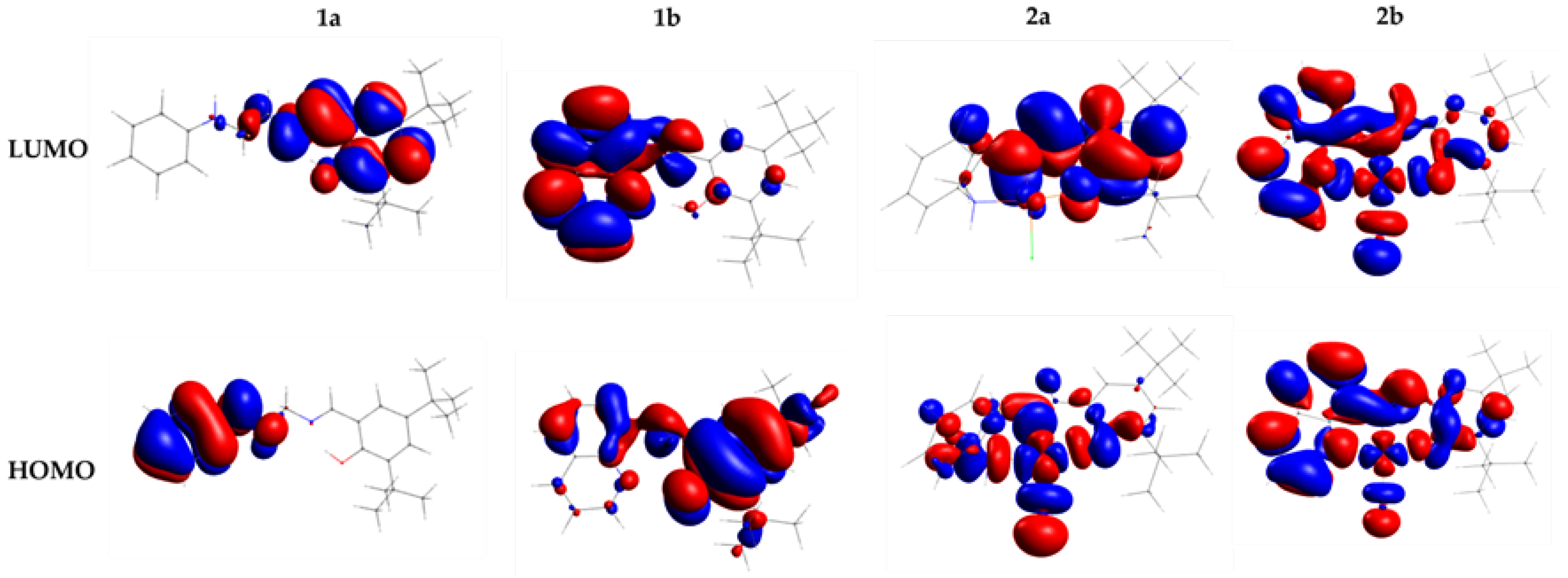
3.1.4. Frontier Molecular Orbital Analysis
3.1.5. Electronic Properties
3.1.6. Studies on the Stability of the Complexes in Solution
3.1.7. Electrochemical Studies
3.2. Biological Activities Studies
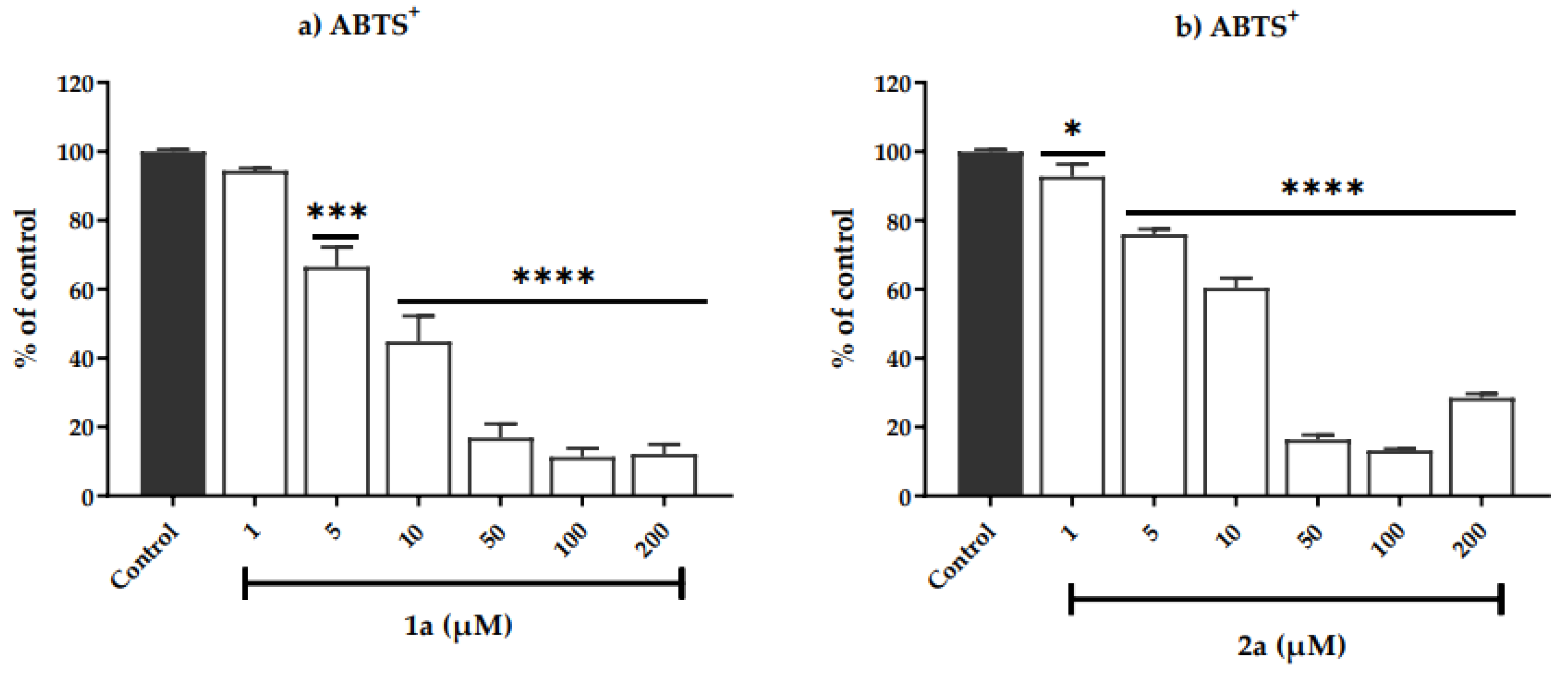
3.2.1. Determination of Antioxidant Capacity
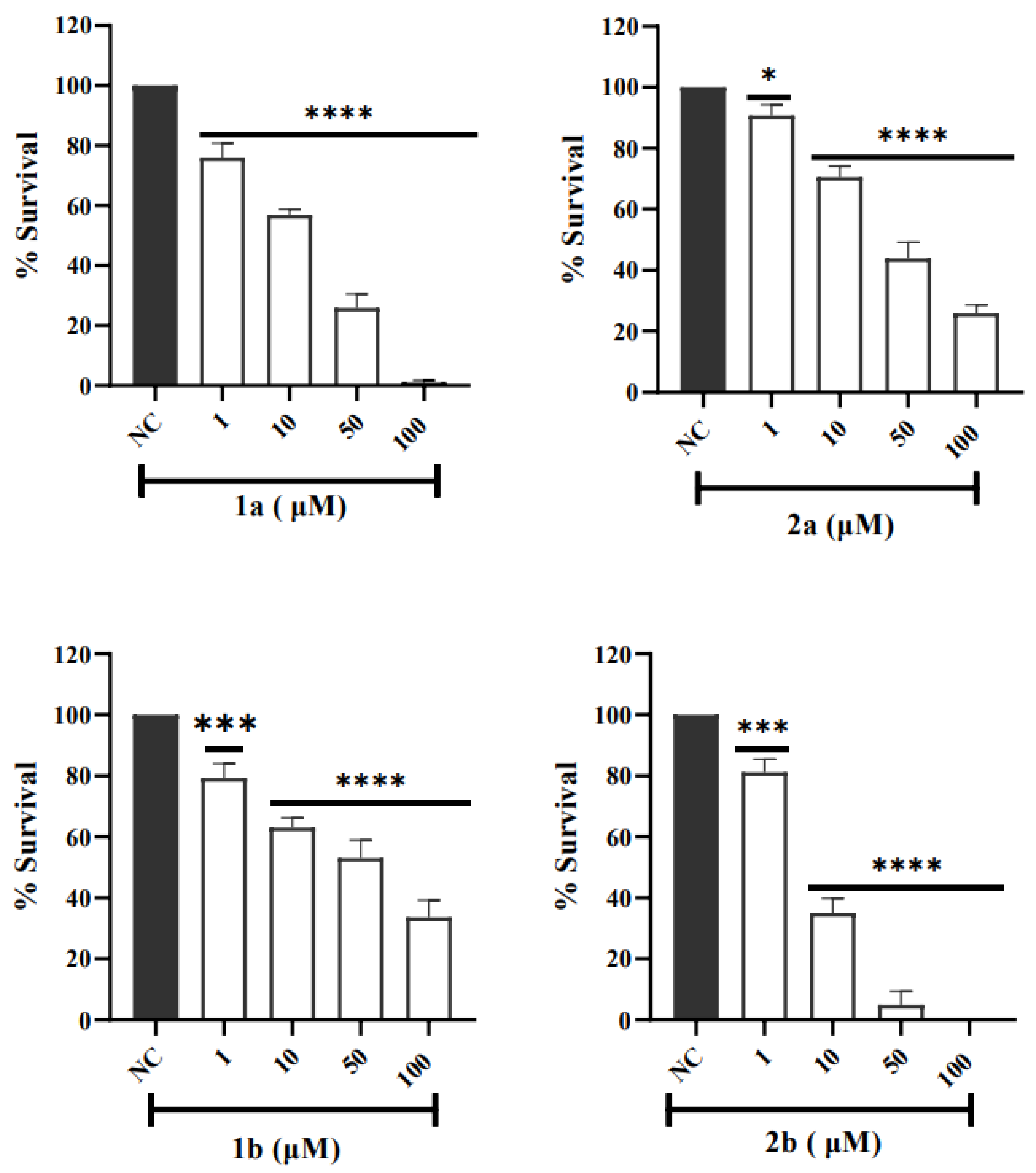
3.2.2. Cytotoxicity Tests in V79 Cells Assessment by MTT Assay
3.2.3. Study of Cytotoxicity in Human Cell Lines
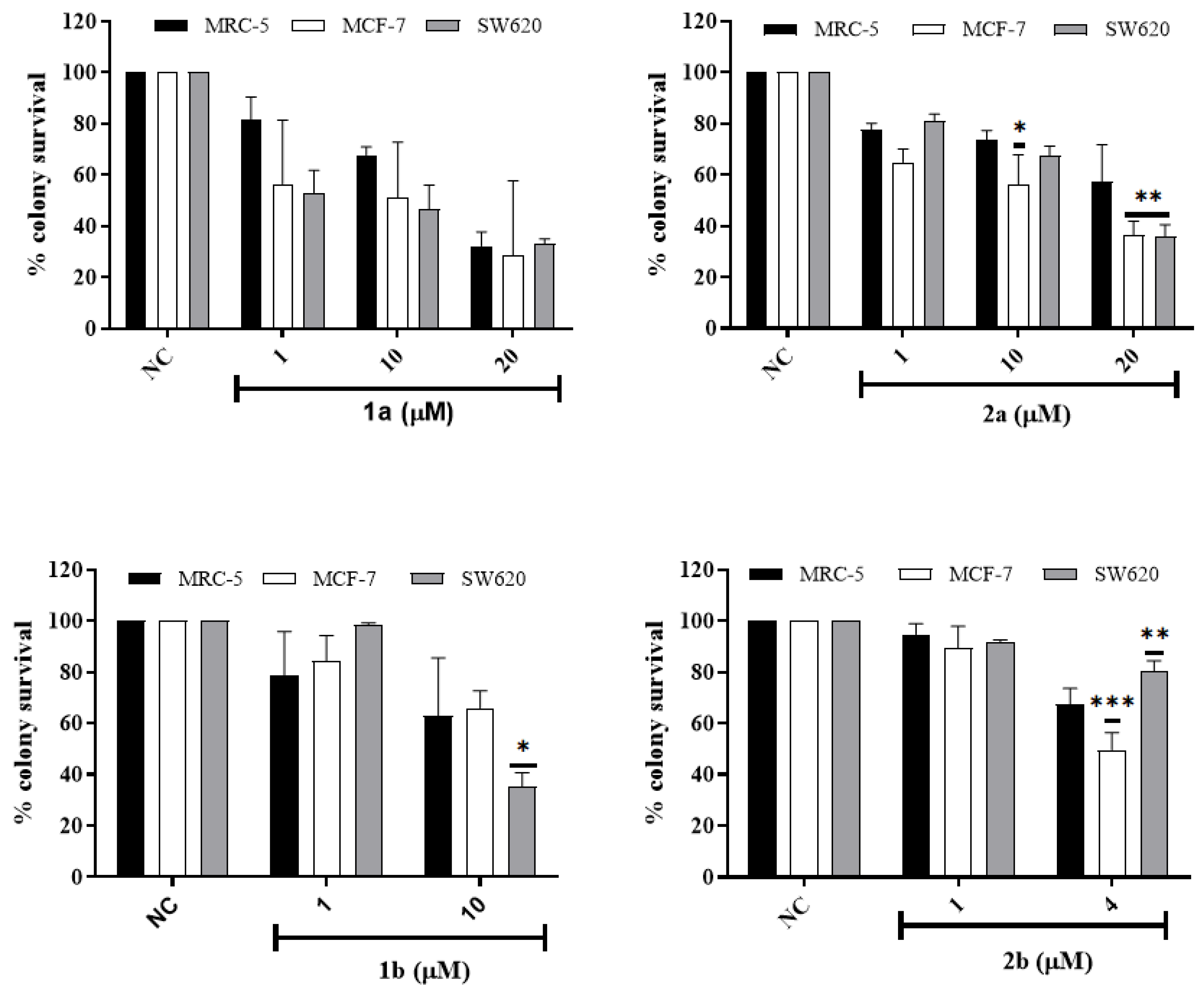
3.2.4. In Vitro Colony Survival Assay
3.2.5. Genotoxicity Effects in Human Cell Lines
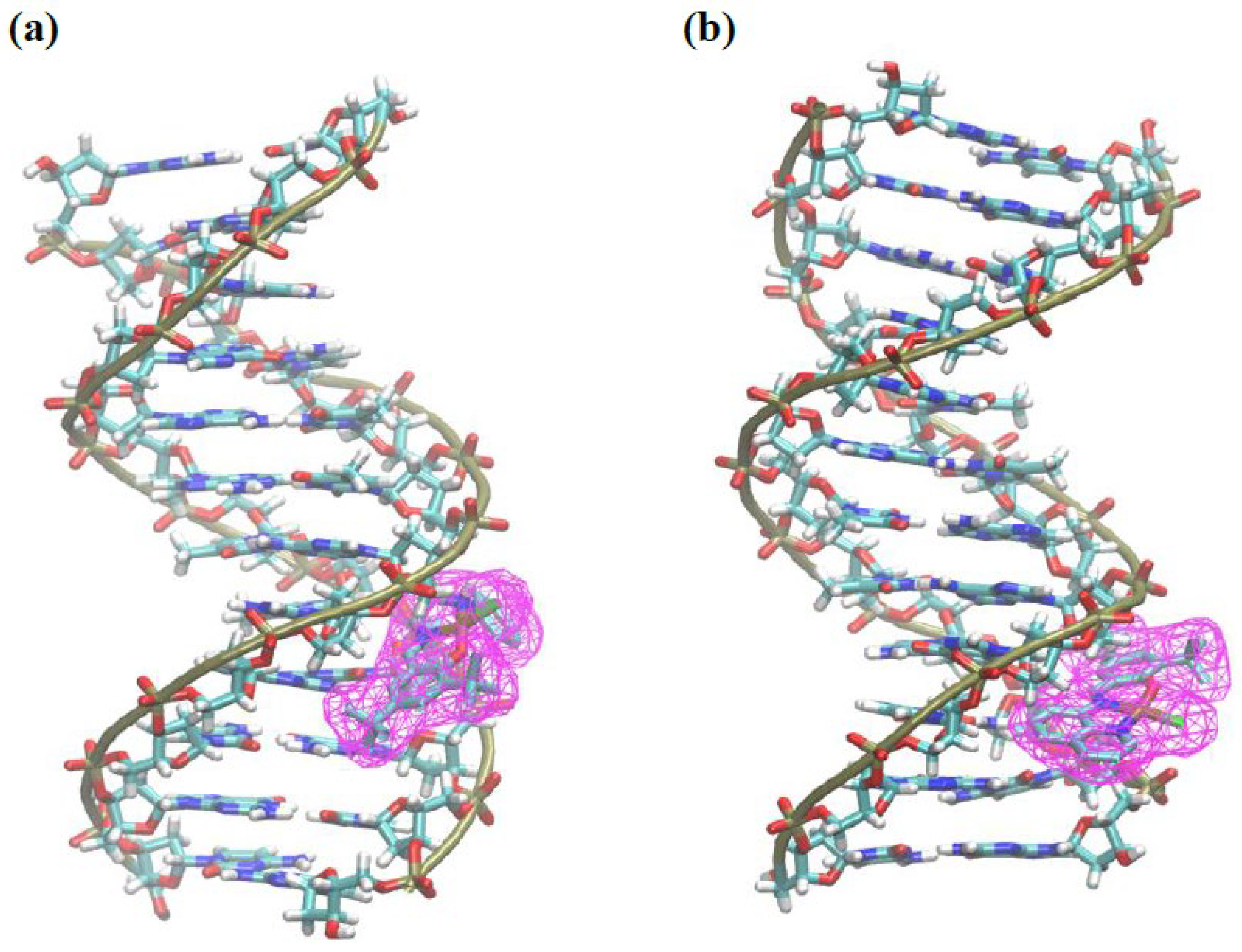
3.2.6. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lucaciu, R.L.; Hangan, A.C.; Sevastre, B.; Oprean, L.S. Metallo-Drugs in Cancer Therapy: Past, Present and Future. Molecules 2022, 27, 6485. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Northcote-Smith, J.; Suntharalingam, K. Emerging Metallopharmaceuticals for the Treatment of Cancer. Trends Chem. 2021, 3, 47–58. [Google Scholar] [CrossRef]
- Yue, S.; Luo, M.; Liu, H.; Wei, S. Recent Advances of Gold Compounds in Anticancer Immunity. Front. Chem. 2020, 8, 2296–2646. [Google Scholar] [CrossRef] [PubMed]
- Lazarević, T.; Rilak, A.; Bugarčić, Ž.D. Platinum, palladium, gold and ruthenium complexes as anticancer agents: Current clinical uses, cytotoxicity studies and future perspectives. Eur. J. Med. Chem. 2017, 142, 8–31. [Google Scholar] [CrossRef]
- Boros, E.; Dyson, P.J.; Gasser, G. Classification of Metal-Based Drugs according to Their Mechanisms of Action. Chem 2020, 6, 41–60. [Google Scholar] [CrossRef]
- Zaki, M.; Arjmand, F.; Tabassum, S. Current and potential of metallo drugs: Revisiting DNA-binding of metal containing molecules and their diverse mechanism of action. Inorg. Chim. Acta 2016, 444, 1–22. [Google Scholar] [CrossRef]
- Arjmand, F.; Afsan, Z.; Sharma, S.; Parveen, S.; Yousuf, I.; Sartaj, S.; Siddique, H.R.; Tabassum, S. Recent advances in metallodrug-like molecules targeting non-coding RNAs in cancer chemotherapy. Coord. Chem. Rev. 2019, 387, 47–59. [Google Scholar] [CrossRef]
- Liang, J.-X.; Zhong, H.-J.; Yang, G.; Vellaisamy, K.; Ma, D.-L.; Leung, C.-H. Recent development of transition metal complexes with In Vivo antitumor activity. J. Inorg. Biochem. 2017, 177, 276–286. [Google Scholar] [CrossRef]
- Rosenberg, B.; VanCamp, L.; Trosko, J.E.; Mansour, V.H. Platinum Compounds: A New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Deo, K.M.; Ang, D.L.; McGhie, B.; Rajamanickam, A.; Dhiman, A.; Khoury, A.; Holland, J.; Bjelosevic, A.; Pages, B.; Gordon, C.; et al. Platinum coordination compounds with potent anticancer activity. Coord. Chem. Rev. 2018, 375, 148–163. [Google Scholar] [CrossRef]
- Gatti, L.; Cassinelli, G.; Zaffaroni, N.; Lanzi, C.; Perego, P. New mechanisms for old drugs: Insights into DNA-unrelated effects of platinum compounds and drug resistance determinants. Drug Resist. Updates 2015, 20, 1–11. [Google Scholar] [CrossRef]
- Fennell, D.A.; Summers, Y.; Cadranel, J.; Benepal, T.; Christoph, D.C.; Lal, R.; Das, M.; Maxwell, F.; Visseren-Grul, C.; Ferry, D. Cisplatin in the modern era: The backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat. Rev. 2016, 44, 42–50. [Google Scholar] [CrossRef]
- Cao, Q.; Li, Y.; Freisinger, E.; Qin, P.Z.; Sigel, R.K.O.; Mao, Z.-W. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs. Inorg. Chem. Front. 2017, 4, 10–32. [Google Scholar] [CrossRef] [Green Version]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Ferraro, M.G.; Piccolo, M.; Misso, G.; Santamaria, R.; Irace, C. Bioactivity and Development of Small Non-Platinum Metal-Based Chemotherapeutics. Pharmaceutics 2022, 14, 954. [Google Scholar] [CrossRef]
- Mahendiran, D.; Amuthakala, S.; Bhuvanesh, N.S.P.; Kumar, R.S.; Rahiman, A.K. Copper complexes as prospective anticancer agents: In vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and S phase arrest. RSC Adv. 2018, 8, 16973–16990. [Google Scholar] [CrossRef] [Green Version]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical pathways of copper complexes: Progress over the past 5 years. Drug Discov. 2021, 26, 1086–1096. [Google Scholar] [CrossRef]
- De Luca, A.; Barile, A.; Arciello, M.; Rossi, L. Copper homeostasis as target of both consolidated and innovative strategies of anti-tumor therapy. J. Trace Elem. Med. Biol. 2019, 55, 204–213. [Google Scholar] [CrossRef]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: ′Copper That Cancer′. Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef]
- Gupte, A.; Mumper, R.J. Elevated Copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009, 35, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Huo, Z.; Qi, X.; Zuo, T.; Wu, Z. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine 2022, 17, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Shagufta; Ahmad, I. Transition metal complexes as proteasome inhibitors for cancer treatment. Inorg. Chim. Acta 2020, 506, 119521. [Google Scholar] [CrossRef]
- Chen, X.; Yang, Q.; Xiao, L.; Tang, D.; Dou, Q.P.; Liu, J. Metal-based proteasomal deubiquitinase inhibitors as potential anticancer agents. Cancer Metastasis Rev. 2017, 36, 655–668. [Google Scholar] [CrossRef] [Green Version]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Copper Complexes as Anticancer Agents Targeting Topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef]
- Ceramella, J.; Mariconda, A.; Iacopetta, D.; Saturnino, C.; Barbarossa, A.; Caruso, A.; Rosano, C.; Sinicropi, M.S.; Longo, P. From coins to cancer therapy: Gold, silver and copper complexes targeting human topoisomerases. Bioorg. Med. Chem. Lett. 2020, 30, 126905. [Google Scholar] [CrossRef]
- Gaikwad, M.; Konkimalla, V.B.; Salunke-Gawali, S. Metal Complexes as topoisomerase inhibitors. Inorg. Chim. Acta 2022, 542, 121089. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Zhou, F. Cuproptosis: A new form of programmed cell death. Cell. Mol. Immunol. 2022, 19, 867–868. [Google Scholar] [CrossRef]
- Gaál, A.; Orgován, G.; Mihucz, V.G.; Pape, I.; Ingerle, D.; Streli, C.; Szoboszlai, N. Metal transport capabilities of anticancer copper chelators. J. Trace Elem. Med. Biol. 2018, 47, 79–88. [Google Scholar] [CrossRef]
- Oliveri, V. Biomedical applications of copper ionophores. Coord. Chem. Rev. 2020, 422, 213474. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Chang, Y.-L.; Liu, S.-T.; Chen, G.-S.; Lee, S.-P.; Huang, S.-M. Differential Cytotoxicity Mechanisms of Copper Complexed with Disulfiram in Oral Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3711. [Google Scholar] [CrossRef]
- Svahn, N.; Moro, A.J.; Roma-Rodrigues, C.; Puttreddy, R.; Rissanen, K.; Baptista, P.V.; Fernandes, A.R.; Lima, J.C.; Rodríguez, L. The Important Role of the Nuclearity, Rigidity, and Solubility of Phosphane Ligands in the Biological Activity of Gold(I) Complexes. Chem. Eur. J. 2018, 24, 14654–14667. [Google Scholar] [CrossRef] [Green Version]
- More, M.S.; Joshi, P.G.; Mishra, Y.K.; Khanna, P.K. Metal complexes driven from Schiff bases and semicarbazones for biomedical and allied applications: A review. Mater. Today Chem. 2019, 14, 100195. [Google Scholar] [CrossRef]
- Abu-Dief, A.M.; Mohamed, I.M.A. A review on versatile applications of transition metal complexes incorporating Schiff bases. Beni Suef Univ. J. Basic Appl. Sci. 2015, 4, 119–133. [Google Scholar] [CrossRef] [Green Version]
- Ceramella, J.; Iacopetta, D.; Catalano, A.; Cirillo, F.; Lappano, R.; Sinicropi, M.S. A Review on the Antimicrobial Activity of Schiff Bases: Data Collection and Recent Studies. Antibiotics 2022, 11, 191. [Google Scholar] [CrossRef]
- Raczuk, E.; Dmochowska, B.; Samaszko-Fiertek, J.; Madaj, J. Different Schiff Bases—Structure, Importance and Classification. Molecules 2022, 27, 787. [Google Scholar] [CrossRef]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. Medchemcomm 2018, 9, 409–436. [Google Scholar] [CrossRef]
- Topkaya, C.G.; Göktürk, T.; Hökelek, T.; Çetin, E.S.; Kincal, S.; Güp, R. In vitro DNA interaction, topoisomerase I/II inhibition and cytotoxic properties of polymeric copper(II) complex bridged with perchlorate ion containing N4-type Schiff base ligand. J. Mol. Struct. 2022, 1266, 133453. [Google Scholar] [CrossRef]
- Uddin, N.; Rashid, F.; Ali, S.; Tirmizi, S.A.; Ahmad, I.; Zaib, S.; Zubair, M.; Diaconescu, P.L.; Tahir, M.N.; Iqbal, J.; et al. Synthesis, characterization, and anticancer activity of Schiff bases. J. Biomol. Struct. Dyn. 2020, 38, 3246–3259. [Google Scholar] [CrossRef]
- Hussain, A.; AlAjmi, M.F.; Rehman, M.T.; Amir, S.; Husain, F.M.; Alsalme, A.; Siddiqui, M.A.; Alkhedhairy, A.A.; Khan, R.A. Copper(II) complexes as potential anticancer and nonsteroidal anti-inflammatory agents: In vitro and in vivo studies. Sci. Rep. 2019, 9, 5237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, Y.P.; Patel, S.K. Molecular structures, spectral, electrochemical, DFT and antioxidant activities of copper(II) complexes with NNO donor Schiff base ligand. J. Mol. Struct. 2021, 1228, 129457. [Google Scholar] [CrossRef]
- Gur′eva, Y.A.; Zalevskaya, O.A.; Shevchenko, O.G.; Slepukhin, P.A.; Makarov, V.A.; Kuchin, A.V. Copper(II) complexes with terpene derivatives of ethylenediamine: Synthesis, and antibacterial, antifungal and antioxidant activity. RCS Adv. 2022, 12, 8841–8851. [Google Scholar] [CrossRef] [PubMed]
- Alothman, A.A.; Al-Farraj, E.S.; Al-Onazi, W.A.; Almarhoon, Z.M.; Al-Mohaimeed, A.M. Spectral characterization, electrochemical, antimicrobial and cytotoxic studies on new metal(II) complexes containing N2O4 donor hexadentate Schiff base ligand. Arab. J. Chem. 2020, 13, 3889–3902. [Google Scholar] [CrossRef]
- Singh, K.; Bhori, M.; Kasu, Y.A.; Bhat, G.; Marar, T. Antioxidants as precision weapons in war against cancer chemotherapy induced toxicity—Exploring the armoury of obscurity. Saudi Pharm. J. 2018, 26, 177–190. [Google Scholar] [CrossRef]
- Afzal, O.; Kumar, S.; Haider, M.R.; Ali, M.R.; Kumar, R.; Jaggi, M.; Bawa, S. A review on anticancer potential of bioactive heterocycle quinoline. Eur. J. Med. Chem. 2015, 97, 871–910. [Google Scholar] [CrossRef]
- Ilakiyalakshmi, M.; Arumugam Napoleon, A.A. Review on recent development of quinoline for anticancer activities. Arab. J. Chem. 2022, 15, 104168. [Google Scholar] [CrossRef]
- Van de Walle, T.; Cools, L.; Mangelinckx, S.; D′hooghe, M. Recent contributions of quinolines to antimalarial and anticancer drug discovery research. Eur. J. Med. Chem. 2021, 226, 113865. [Google Scholar] [CrossRef]
- Gomes, F.S.; Bergamo, A.L.; Casagrande, O.L. Synthesis and Characterization of IminoPhenolate Titanium Complexes and Their Use in Homo- and Copolymerization of Ethylene. Macromol. Chem. Phys. 2014, 215, 1735–1743. [Google Scholar] [CrossRef]
- Bouyhayi, M.; Sarazin, Y.; Osvaldo, L.; Casagrande, O.L.; Carpentier, J.-F. Aluminum, calcium and zinc complexes supported by potentially tridentate iminophenolate ligands: Synthesis and use in the ring-opening polymerization of lactide. Appl. Organomet. Chem. 2012, 26, 681–688. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Brandenburg, K.; Putz, H. Diamond—Crystal and Molecular Structure Visualization; Crystal Impact GbR: Bonn, Germany, 2006. [Google Scholar]
- Neese, F. Sofware update: The ORCA program system, version 4.0. WIREs Comput. Mol. Sci. 2017, 8, e1327. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter. Mater. Phys. 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Schäefer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef] [Green Version]
- Weigend, F.; Ahlrichs, R. Balanced basis set of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Reger, D.L.; Pascui, A.E.; Foley, E.A.; Smith, M.D.; Jezierska, J.; Wojciechowska, A.; Stoian, S.A.; Ozarowski, A. Dinuclear Metallacycles with Single M−X−M Bridges (X = Cl−, Br−; M= Fe(II), Co(II), Ni(II), Cu(II), Zn(II), Cd(II)): Strong Antiferromagnetic Superexchange Interactions. Inorg. Chem. 2017, 56, 2884–2901. [Google Scholar] [CrossRef]
- Weigend, F. Accurate Coulomb-Fitting Basis Set for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Hansen, A.; Becker, U. Efficient, approximate and parallel Hartree-Fock and hybrid DFT calculations. A ′chain-of-spheres′ algorithm for the Hartree-Fock exchange. Chem. Phys. 2009, 356, 98–109. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acid Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Biovia, D.S. Discovery Studio Modeling Environment; Release 2017; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Sevgi, K.; Tepe, B.; Sarikurkcu, C. Antioxidant and DNA damage protection potentials of selected phenolic acids. Food Chem. Toxicol. 2015, 77, 12–21. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Iftikhar, B.; Javed, K.; Khan, M.S.U.; Akhter, Z.; Mirza, B.; Mckee, V. Synthesis, characterization and biological assay of Salicylaldehyde Schiff base Cu(II) complexes and their precursors. J. Mol. Struct. 2018, 1155, 337–348. [Google Scholar] [CrossRef]
- Sonmez, F.; Gunesli, Z.; Kurt, B.Z.; Gazioglu, I.; Avci, D.; Kucukislamoglu, M. Synthesis, antioxidant activity and SAR study of novel spiro-isatin-based Schiff bases. Mol. Divers. 2019, 23, 829–844. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Azqueta, A.; Langie, S.A.S.; Boutet-Robinet, E.; Duthie, S.; Ladeira, C.; Møller, P.; Collins, A.R.; Godschalk, R.W.L. DNA repair as a human biomonitoring tool: Comet assay approaches. Mutat. Res. Rev. Mutat. Res. 2019, 781, 71–87. [Google Scholar] [CrossRef]
- Abo-Aly, M.M.; Salem, A.M.; Sayed, M.A.; Abdel Aziz, A.A. Spectroscopic and structural studies of the Schiff base 3-methoxy-N-salicylidene-o-amino phenol complexes with some transition metal ions and their antibacterial, antifungal activities. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 136, 993–1000. [Google Scholar] [CrossRef]
- Keškić, T.; Čobeljić, B.; Gruden, M.; Anđelković, K.; Pevec, A.; Turel, I.; Radanović, D.; Zlatar, M. What Is the Nature of Interactions of BF4−, NO3−, and ClO− to Cu(II) Complexes with Girard′s T Hydrazine? When Can Binuclear Complexes Be Formed? Cryst. Growth Des. 2019, 19, 4810–4821. [Google Scholar] [CrossRef]
- Fernandes, C.; Horn, A.; Da Motta, O.V.; De Assis, V.M.; Rocha, M.R.; Mathias, L.S.; Bull, E.S.; Bortoluzzi, A.J.; Guimarães, E.V.; Almeida, J.C.A.; et al. Synthesis, characterization and antibacterial activity of FeIII, CoII, CuII and ZnII complexes probed by transmission electron microscopy. J. Inorg. Biochem. 2010, 104, 1214–1223. [Google Scholar] [CrossRef]
- De Oliveira Neto, J.G.; Filho, J.G.S.; Bittar, E.M.; Silva, L.M.; Sousa, F.F.; Domingos, H.V.; Costa-Lotufo, A.L.; Reis, V.S.; dos Santos, A.O. Structural, thermal, electronic, vibrational, magnetic, and cytotoxic properties of chloro(glycinato-N,O)(1,10-phenanthroline-N,N′)-copper(II) trihydrate coordination complex. J. Inorg. Biochem. 2022, 226, 111658–111666. [Google Scholar] [CrossRef]
- Santiago, P.H.O.; Tiago, F.S.; Castro, M.S.; Souza, P.E.N.; Martins, J.B.L.; Gatto, C.C. DFT analysis, spectroscopic study and biological activity of a newly synthesized benzoylhydrazone binuclear Cu(II) complex. J. Inorg. Biochem. 2020, 204, 110949–110957. [Google Scholar] [CrossRef]
- Sasmal, A.; Shit, S.; Rizzoli, C.; Wang, H.; Desplanches, C.; Mitra, S. Framework Solids Based on Copper(II) Halides (Cl/Br) and Methylene-Bridged Bis(1-hydroxybenzotriazole): Synthesis, Crystal Structures, Magneto-Structural Correlation, and Density Functional Theory (DFT) Studies. Inorg. Chem. 2012, 51, 10148–10157. [Google Scholar] [CrossRef]
- Kerflani, A.; Si Larbi, K.S.; Rabahi, A.; Bouchoucha, A.; Zaater, S.; Terrachet-Bouaziz, S. Novel palladium (II) complexes with iminocoumarin ligands: Synthesis, characterisation, electrochemical behaviour, DFT calculations and biological activities, ADMET study and molecular docking. Inorg. Chim. Acta 2022, 529, 120659. [Google Scholar] [CrossRef]
- Choudhary, V.; Bhatt, A.; Dash, D.; Sharma, N. DFT calculations on molecular structures, HOMO–LUMO study, reactivity descriptors and spectral analyses of newly synthesized diorganotin(IV) 2-chloridophenylacetohydroxamate complexes. J. Comput. Chem. 2019, 40, 2359–2363. [Google Scholar] [CrossRef]
- Mamta; Subhash; Pinki; Chaudhary, A. In vitro cytotoxicity and antimicrobial evaluation of novel 24–28 membered Schiff base octaazamacrocyclic complexes of manganese(II): Synthesis, characterization, DFT and molecular docking studies. J. Mol. Struct. 2023, 1275, 134667. [Google Scholar] [CrossRef]
- Zolezzi, S.; Spodine, E.; Decinti, A. Electrochemical studies of copper(II) complexes with Schiff-base ligands. Polyhedron 2002, 21, 55–59. [Google Scholar] [CrossRef]
- Saghatforoush, L.A.; Hosseinpour, S.; Bezpalko, M.W.; Kassel, W.S. X-ray crystal structural and spectral studies of copper(II) and nickel(II) complexes of functionalized bis(thiosemicarbazone) ligands and investigation of their electrochemical behavior. Inorg. Chim. Acta 2019, 484, 527–534. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative Stress: Harms and Benefits for Human Health. Oxid. Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef] [Green Version]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Saeidnia, S.; Abdollahi, M. Antioxidants: Friends or foe in prevention or treatment of cancer: The debate of the century. Toxicol. Appl. Pharmacol. 2013, 271, 49–63. [Google Scholar] [CrossRef]
- Dastmalchi, N.; Baradaran, B.; Latifi-Navid, S.; Safaralizadeh, R.; Khojasteh, S.M.B.; Amini, M.; Roshani, E.; Lotfinejad, P. Antioxidants with two faces toward cancer. Life Sci. 2020, 258, 118186. [Google Scholar] [CrossRef]
- Shah, S.S.; Shah, D.; Khan, I.; Ahmad, S.; Ali, U.; Rahman, A. Synthesis and Antioxidant Activities of Schiff Bases and Their Complexes: An Updated Review. Biointerface Res. Appl. Chem. 2020, 10, 6936–6963. [Google Scholar] [CrossRef]
- Marchi, R.C.; Campos, I.A.S.; Santana, V.T.; Carlos, R.M. Chemical implications and considerations on techniques used to assess the in vitro antioxidant activity of coordination compounds. Coord. Chem. Rev. 2022, 451, 214275. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant Determinations by the Use of a Stable Free Radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Asgarshamsi, M.H.; Fassihi, A.; Hassanzadeh, F.; Saghaei, L.; Attar, A.M.; Mohammad-Beigi, H. Synthesis, antioxidant activity, and density functional theory study of some novel 4-[(benzo[d]thiazol-2-ylimino)methyl]phenol derivatives: A comparative approach for the explanation of their radical scavenging activities. Res. Pharm. Sci. 2021, 16, 35–47. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Taie, H.A.A.; Bakheit, A.H.; Mostafa, G.A.E.; Marzouk, M.; Rashid, H.; Al-Salahi, R. Investigation of 4-Hydrazinobenzoic Acid Derivatives for Their Antioxidant Activity: In Vitro Screening and DFT Study. ACS Omega 2021, 6, 31993–32004. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Haun, M. Cytotoxicity of trans-Dehydrocrotonin from Croton cajucara on V79 Cells and Rat Hepatocytes. Planta Med. 1999, 65, 522–526. [Google Scholar] [CrossRef]
- Sharma, R.I.; Smith, T.A.D. Colorectal Tumor Cells Treated with 5-FU, Oxaliplatin, Irinotecan, and Cetuximab Exhibit Changes in 18F-FDG Incorporation Corresponding to Hexokinase Activity and Glucose Transport. J. Nucl. Med. 2008, 49, 1386–1394. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Wei, K.; Gao, Y.; Zhou, Z.; Zheng, X.; Li, J.; Qi, J. Comparative evaluation of the structure and antitumor mechanism of mononuclear and trinucleated thiosemicarbazone Cu(II) complexes. J. Inorg. Biochem. 2023, 240, 112116. [Google Scholar] [CrossRef]
- Kabakov, A.E.; Gabai, V.L. Cell Death and Survival Assays. Methods Mol. Biol. 2018, 1709, 107–127. [Google Scholar] [CrossRef]
- Pich, C.T.; Dos Santos, P.R.; Fortunato, T.V.O.; Chiarello, M.; De Oliveira, I.M.; Soares, B.Q.; Ghermani, N.E.; Machado, M.; Roesch-Ely, M.; Dumas, F.; et al. Mixed Ternary Mononuclear Copper(II) Complexes Based on Valproic Acid with 1,10-Phenanthroline and 2,2′-Bipyridine Ligands: DNA Interaction and Cytotoxicity in V79 Cells. J. Braz. Chem. Soc. 2019, 30, 597–613. [Google Scholar] [CrossRef]
- Erxleben, A. Interactions of copper complexes with nucleic acids. Coord. Chem. Rev. 2018, 360, 92–121. [Google Scholar] [CrossRef]


| 2a | |||
|---|---|---|---|
| Bond Lengths (Å) | |||
| Cu(1)-O(1) | 1.904(13) | Cu(2)-O(2) | 1.876(13) |
| Cu(1)-N(2) | 1.922(16) | Cu(2)-N(4) | 1.932(16) |
| Cu(1)-N(1) | 2.190(16) | Cu(2)-N(3) | 2.023(16) |
| Cu(1)-Cl(1) | 2.250(5) | Cu(2)-Cl(2) | 2.245(5) |
| Cu(1)-Cl(1)#1 | 2.845(6) | ||
| Bond Angles (°) | |||
| O(1)-Cu(1)-N(2) | 92.97(6) | O(2)-Cu(2)-N(4) | 92.45(6) |
| O(1)-Cu(1)-N(1) | 174.53(7) | O(2)-Cu(2)-N(3) | 171.51(7) |
| N(2)-Cu(1)-N(1) | 84.59(6) | N(4)-Cu(2)-N(3) | 84.68(7) |
| O(1)-Cu(1)-Cl(1) | 91.03(4) | O(2)-Cu(2)-Cl(2) | 91.99(4) |
| N(2)-Cu(1)-Cl(1) | 174.79(5) | N(4)-Cu(2)-Cl(2) | 172.94(5) |
| N(1)-Cu(1)-Cl(1) | 91.13(5) | N(3)-Cu(2)-Cl(2) | 91.67(5) |
| Cl(1)-Cu(1)-Cl(1)#1 | 89.48(17) | ||
| Energy (eV) | 1a | 1b | 2a | 2b |
|---|---|---|---|---|
| EHOMO | −5.442 | −5.697 | −5.392 | −5.477 |
| ELUMO | −1.465 | −1.939 | −2.594 | −2.766 |
| ∆E = ELUMO − EHOMO | 3.977 | 3.758 | 2.798 | 2.711 |
| Physicochemical Parameters | Mathematical Relationship | Calculated with B3LYP/def2-TZVPP End def2-SVP | |||
|---|---|---|---|---|---|
| Energy (eV) or a eV−1 | - | 1a | 1b | 2a | 2b |
| η | η | 1.988 | 1.879 | 1.399 | 1.355 |
| ς | ς | 0.251 a | 0.266 a | 0.357 a | 0.369 a |
| IP | −EHOMO | 5.442 | 5.697 | 5.392 | 5.477 |
| EA | −ELUMO | 1.465 | 1.939 | 2.594 | 2.766 |
| μ | μ | −3.453 | −3.818 | −3.993 | −4.121 |
| χ | χ | 3.453 | 3.818 | 3.993 | 4.121 |
| ϖ | ϖ | 2.999 | 3.879 | 5.698 | 6.264 |
| Compound | Epc/V | Epa/V | E1/2/V | ΔE/mV |
|---|---|---|---|---|
| 1a | - | 1.00 1.56 | - | |
| 1b | - | 1.10 1.40 | - | |
| 2a | 0.19 | 0.48 0.94 1.39 | 0.33 | 240 |
| 2b | 0.14 | 0.51 1.12 1.39 | 0.32 | 356 |
| Concentration (μM) | 2a | 2b | ||
|---|---|---|---|---|
| Inhibition (%) | Inhibition (%) | |||
| DPPH• | ABTS+ | DPPH• | ABTS+ | |
| Control | 100.0 ± 0.5 | 100.0 ± 0.6 | 100.0 ± 0.6 | 100.0 ± 0.6 |
| 1 | 96.7 ± 1.3 | 92.7 ± 3.6 * | 96.5 ± 3.6 | 102.0 ± 2.6 |
| 5 | 76.5 ± 7.7 *** | 76.0 ± 1.5 **** | 94.6 ± 1.5 | 102.3 ± 3.1 |
| 10 | 22.19 ± 3.3 **** | 60.4 ± 2.8 **** | 96.0 ± 1.9 | 95.6 ± 0.9 |
| 50 | 13.9 ± 1.3 **** | 16.4 ± 1.4 **** | 92.5 ± 6.6 | 93.1 ± 0.9 |
| 100 | 21.9 ± 1.2 **** | 13.3 ± 0.4 **** | 61.36 ± 4.1 **** | 77.7 ± 0.8 **** |
| 200 | 37.7 ± 2.2 **** | 28.5 ± 1.1 **** | a | 46.7 ± 4.7 **** |
| Imax (%) | 86.1 ± 1.4 | 87.7 ± 0.4 | 38.6 ± 4.1 | 53.3 ± 4.7 |
| IC50 (μM) | 6.3 ± 0.0 | 11.8 ± 0.0 | - | 100.7 ± 0.0 |
| V79 Cell Line | ||||
|---|---|---|---|---|
| Compounds | 1a | 1b | 2a | 2b |
| IC50 (µM) | 23 ± 0 | 69 ± 9 | 19 ± 1 | 8 ± 4 |
| IC50 (µM) | |||
|---|---|---|---|
| Cells | MRC-5 | SW620 | MCF-7 |
| 1a | 14.7 ± 1.1 (24 h) 17.1 ± 3.3 (72 h) | 27.6 ± 1.2 (24 h) 16.4 ± 2.4 (72 h) | 30.5 ± 4.4 (24 h) 28.6 ± 1.6 (72 h) |
| 1b | 63.4 ± 5.5 (24 h) 15.5 ± 9.0 (72 h) | 41.9 ± 9.7 (24 h) 10.5 ± 5.3 (72 h) | 110.6 ± 9.5 (24 h) 44.5 ± 4.3 (72 h) |
| 2a | 53.9 ± 11.0 (24 h) 15.4 ± 4.1 (72 h) | 33.9 ± 3.7 (24 h) 30.0 ± 2.9 (72 h) | 79.9 ± 7.8 (24 h) 25.5 ± 3.8 (72 h) |
| 2b | 11.0 ± 0.1(24 h) 6.1 ± 0.4 (72 h) | 8.3 ± 1.2 (24 h) 4.2 ± 0.3 (72 h) | 3.6 ± 0.6 (24 h) 5.2 ± 0.4 (72 h) |
| Compound | Dose (μM) | MRC-5 | MCF-7 | SW620 |
|---|---|---|---|---|
| Damage Índex (DI) ± SD | ||||
| 1a | 0 | 7.3 ± 3.2 | 7.7 ± 3.5 | 12.7 ± 2.5 |
| 1 | 19.3 ± 3.9 | 14.7 ± 3.2 | 16.7 ± 2.5 | |
| 10 | 39.7 ± 9.5 * | 34.3 ± 4.2 *** | 21.3 ± 6.7 | |
| 50 | 121.3 ± 10.0 **** | 100.7 ± 2.9 **** | 107.0 ± 7.2 **** | |
| 100 | 200.3 ± 22.0 **** | 154.7 ± 7.2 **** | 212.3 ± 2.5 **** | |
| 2a | 0 | 5.7 ± 2.1 | 5.7 ± 2.5 | 7.3 ± 2.5 |
| 1 | 11.0 ± 2.0 | 8.3 ± 2.1 | 23.7 ± 2.1 | |
| 10 | 17.3 ± 3.1 | 15.7 ± 2.9 | 36.0 ± 4.6 ** | |
| 50 | 63.0 ± 6.0 **** | 38.7 ± 3.5 **** | 96.7 ± 11.4 **** | |
| 100 | 111.0 ± 7.8 **** | 77.3 ± 9.0 **** | 104.3 ± 13.6 **** | |
| 1b | 0 | 9.7 ± 3.1 | 8.0 ± 1.0 | 6.7 ± 2.5 |
| 1 | 16.3 ± 2.5 | 16.0 ± 4.0 | 10.0 ± 1.0 | |
| 10 | 20.0 ± 5.2 | 22.3 ± 6.4 | 34.7 ± 7.4 *** | |
| 50 | 101.0 ± 5.0 **** | 52.7 ± 9.5 **** | 120.0 ± 5.3 **** | |
| 100 | 135.0 ± 6.2 **** | 102.0 ± 6.0 **** | 236.0 ± 8.2 **** | |
| 2c | 0 | 5.7 ± 2.5 | 6.7 ± 2.5 | 5.3 ± 1.5 |
| 1 | 10.0 ± 2.7 | 21.3 ± 5.5 | 14.0 ± 5.2 | |
| 10 | 19.0 ± 6.2 | 93.7 ± 6.7 **** | 31.3 ± 5.5 ** | |
| 50 | 75.0 ± 7.8 **** | 252.3 ± 9.5 **** | 109.0 ± 13.1 **** | |
| 100 | 220.3 ± 13.3 **** | 250.3 ± 4.9 **** | 250.7 ± 10.0 **** | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, A.C.; Nunes, I.J.; Ferreira, W.V.; Tomasini, P.P.; Trindade, C.; Martins, C.C.; Wilhelm, E.A.; Oliboni, R.d.S.; Netz, P.A.; Stieler, R.; et al. Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups. Pharmaceutics 2023, 15, 376. https://doi.org/10.3390/pharmaceutics15020376
Pinheiro AC, Nunes IJ, Ferreira WV, Tomasini PP, Trindade C, Martins CC, Wilhelm EA, Oliboni RdS, Netz PA, Stieler R, et al. Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups. Pharmaceutics. 2023; 15(2):376. https://doi.org/10.3390/pharmaceutics15020376
Chicago/Turabian StylePinheiro, Adriana Castro, Ianka Jacondino Nunes, Wesley Vieira Ferreira, Paula Pellenz Tomasini, Cristiano Trindade, Carolina Cristóvão Martins, Ethel Antunes Wilhelm, Robson da Silva Oliboni, Paulo Augusto Netz, Rafael Stieler, and et al. 2023. "Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups" Pharmaceutics 15, no. 2: 376. https://doi.org/10.3390/pharmaceutics15020376