Rational Design of Hydrogels for Cationic Antimicrobial Peptide Delivery: A Molecular Modeling Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. AMP Selection
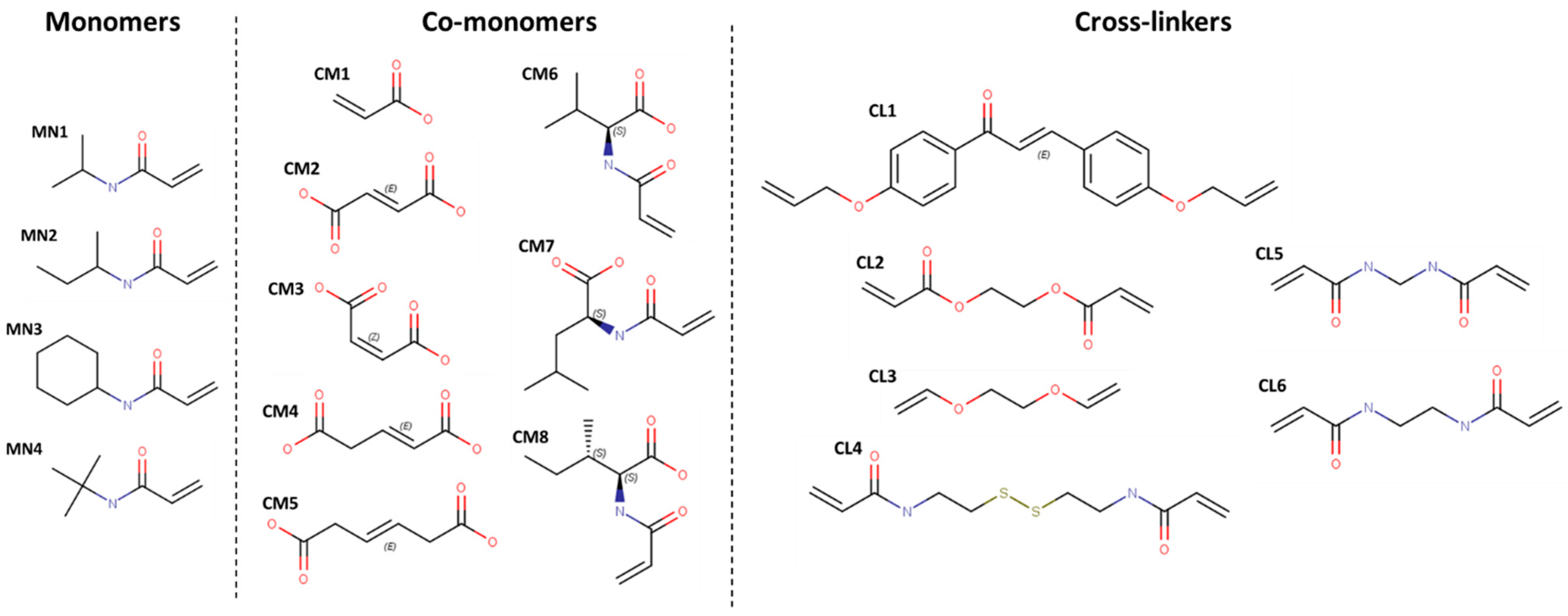
2.2. Hydrogel Building Blocks Selection
2.3. Analysis of the Affinity between Hydrogel Building Blocks and cAMPs
2.4. Ensemble and Simulations of Hydrogel-cAMP Complexes
3. Results and Discussion
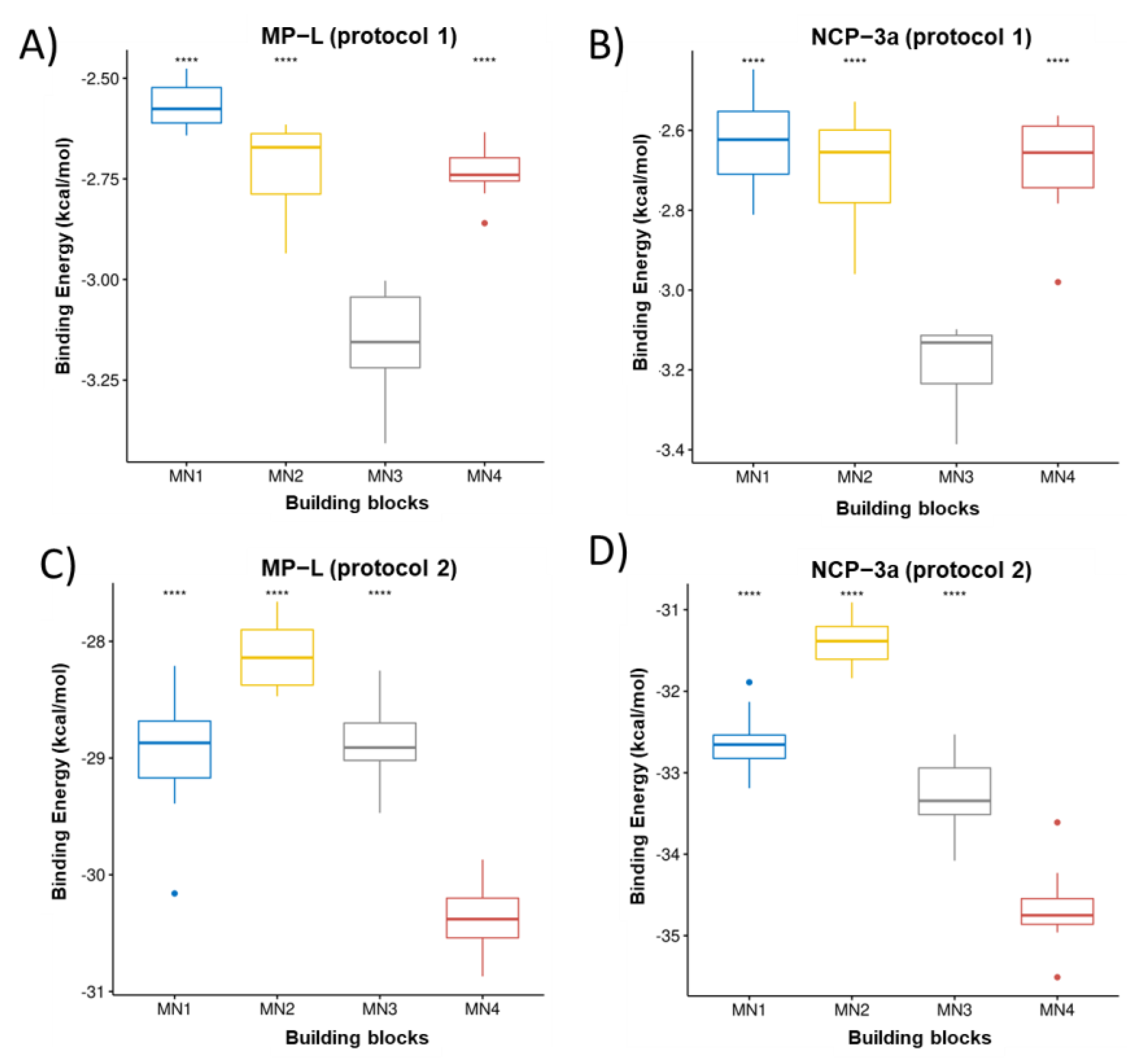
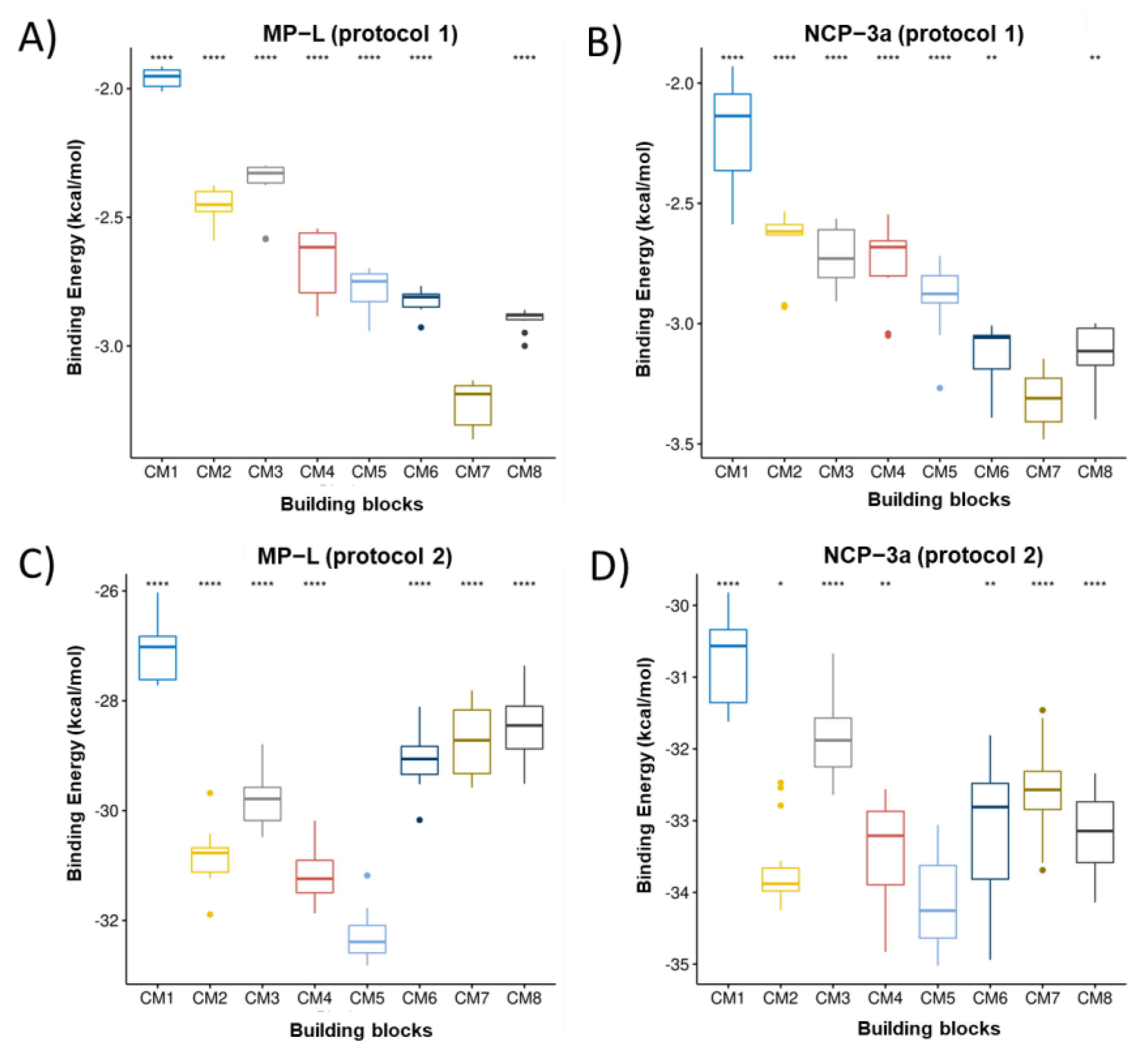
3.1. Ranking of Building Blocks by Molecular Docking
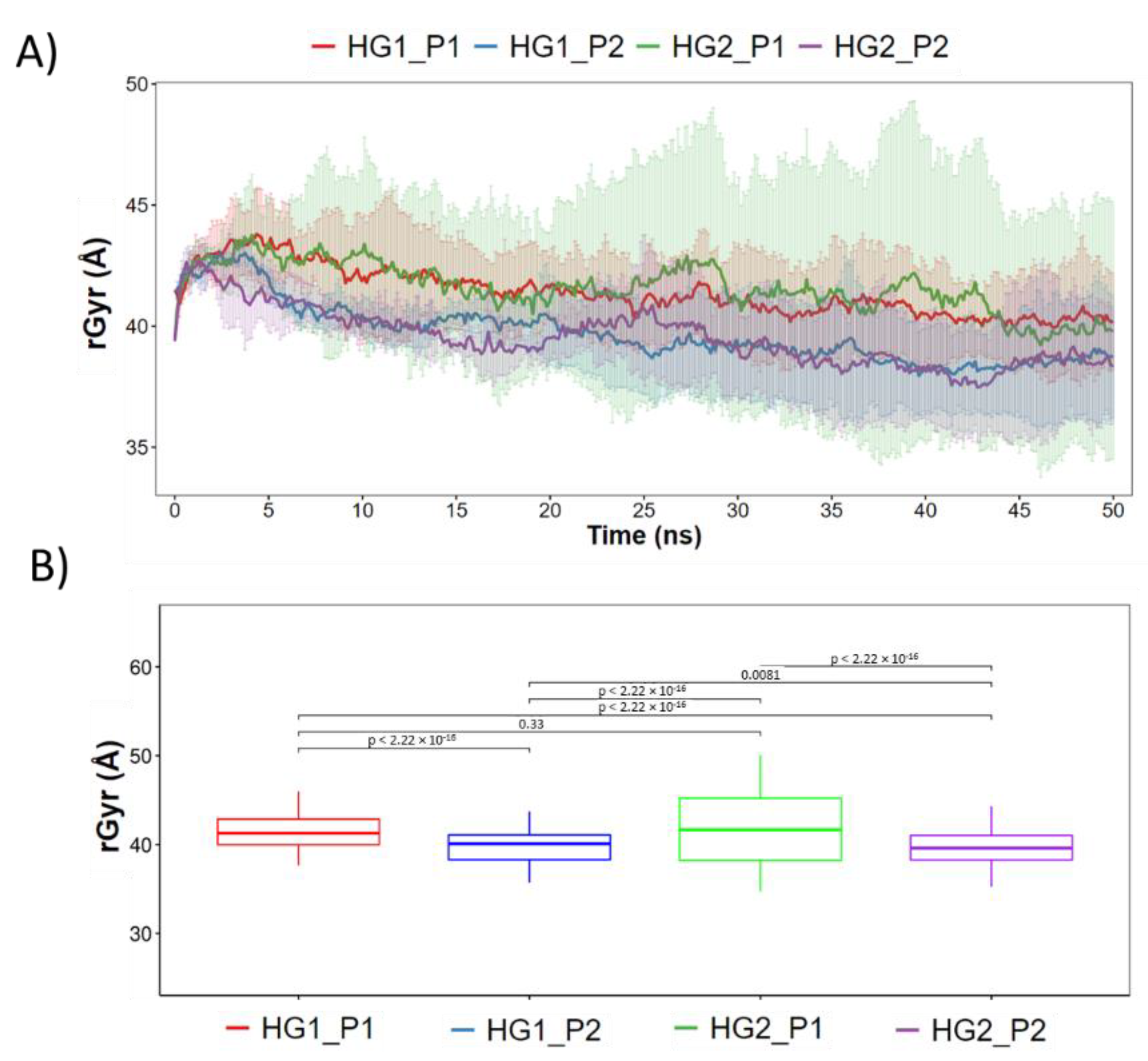
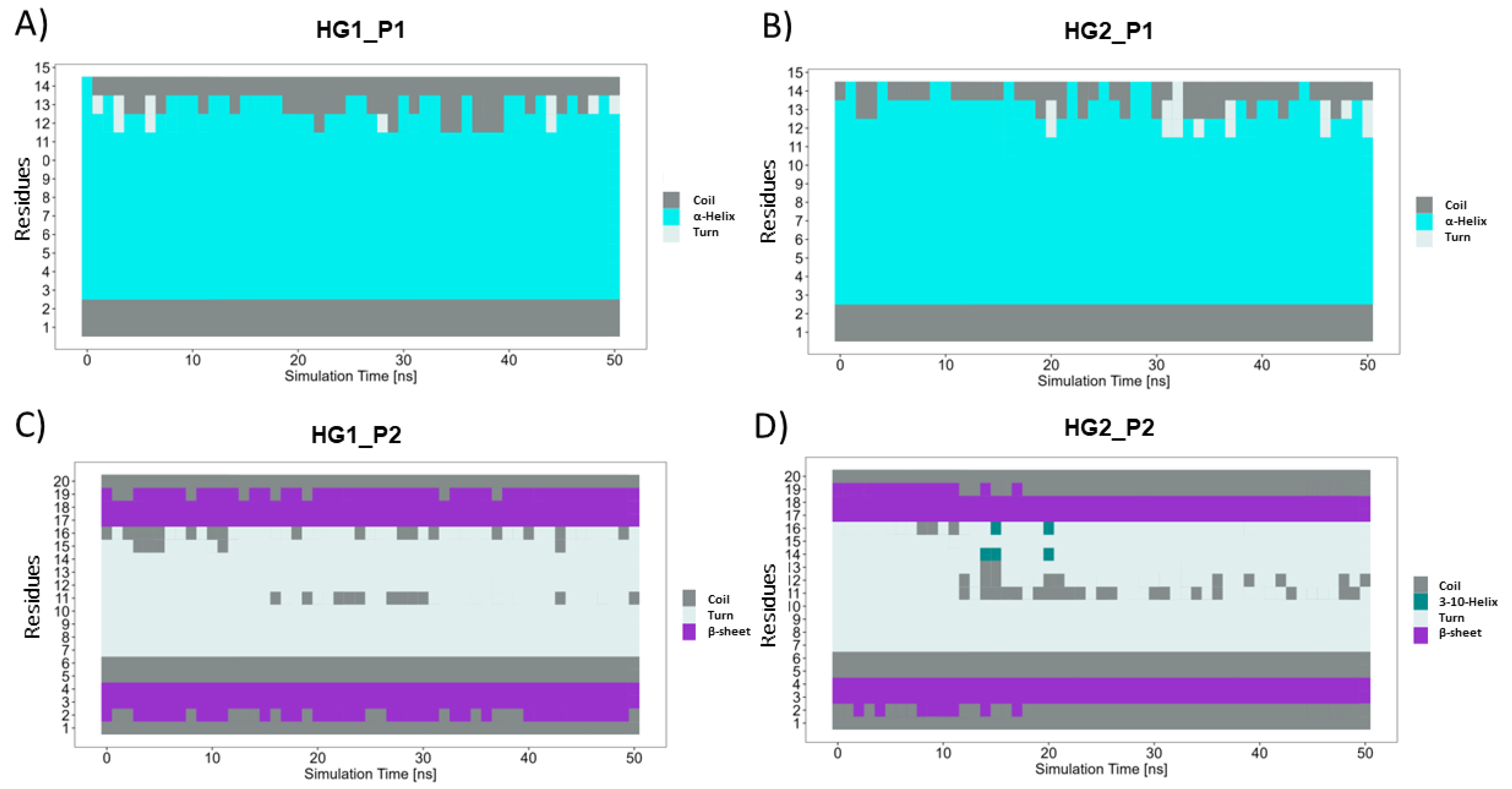
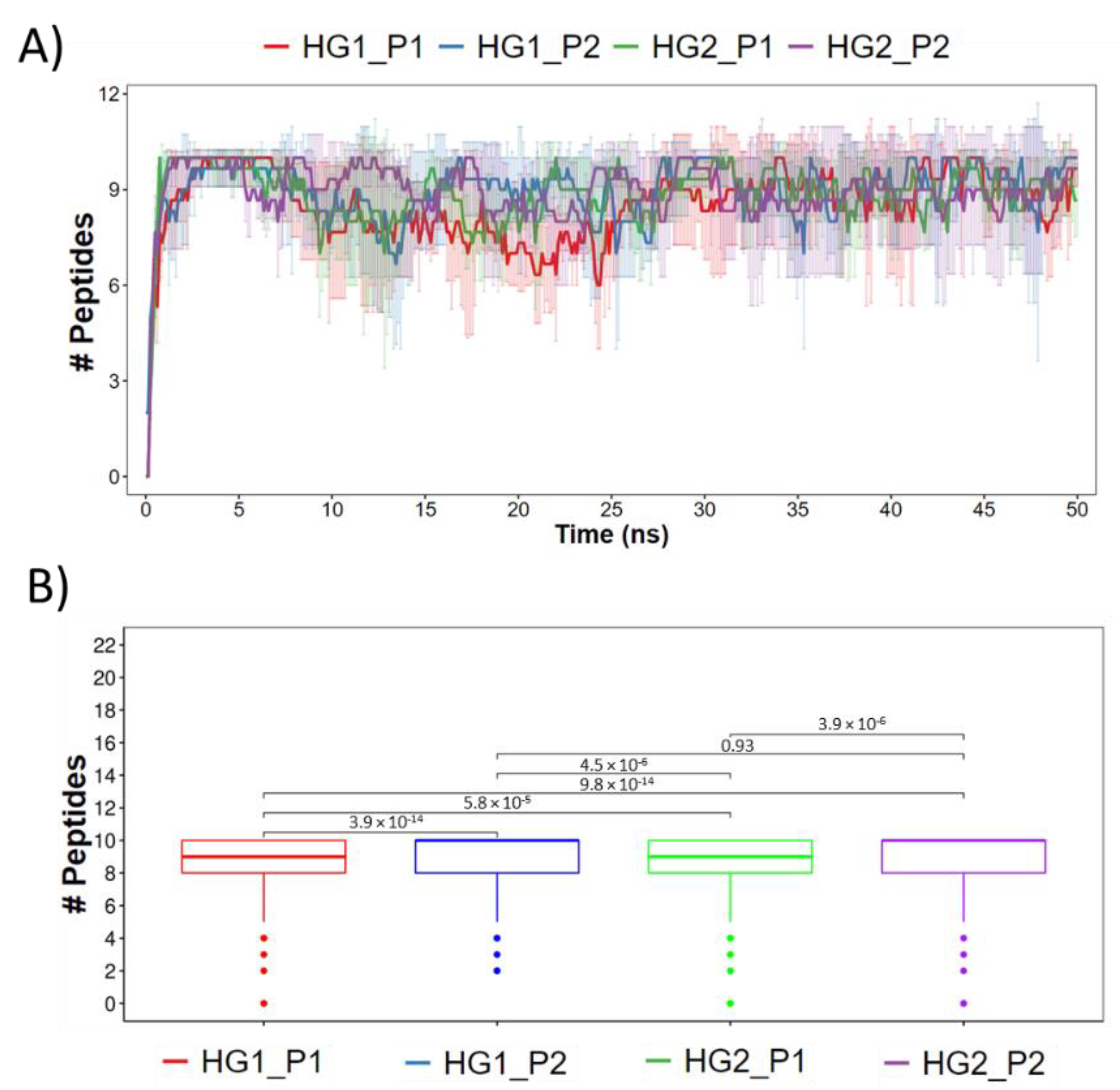
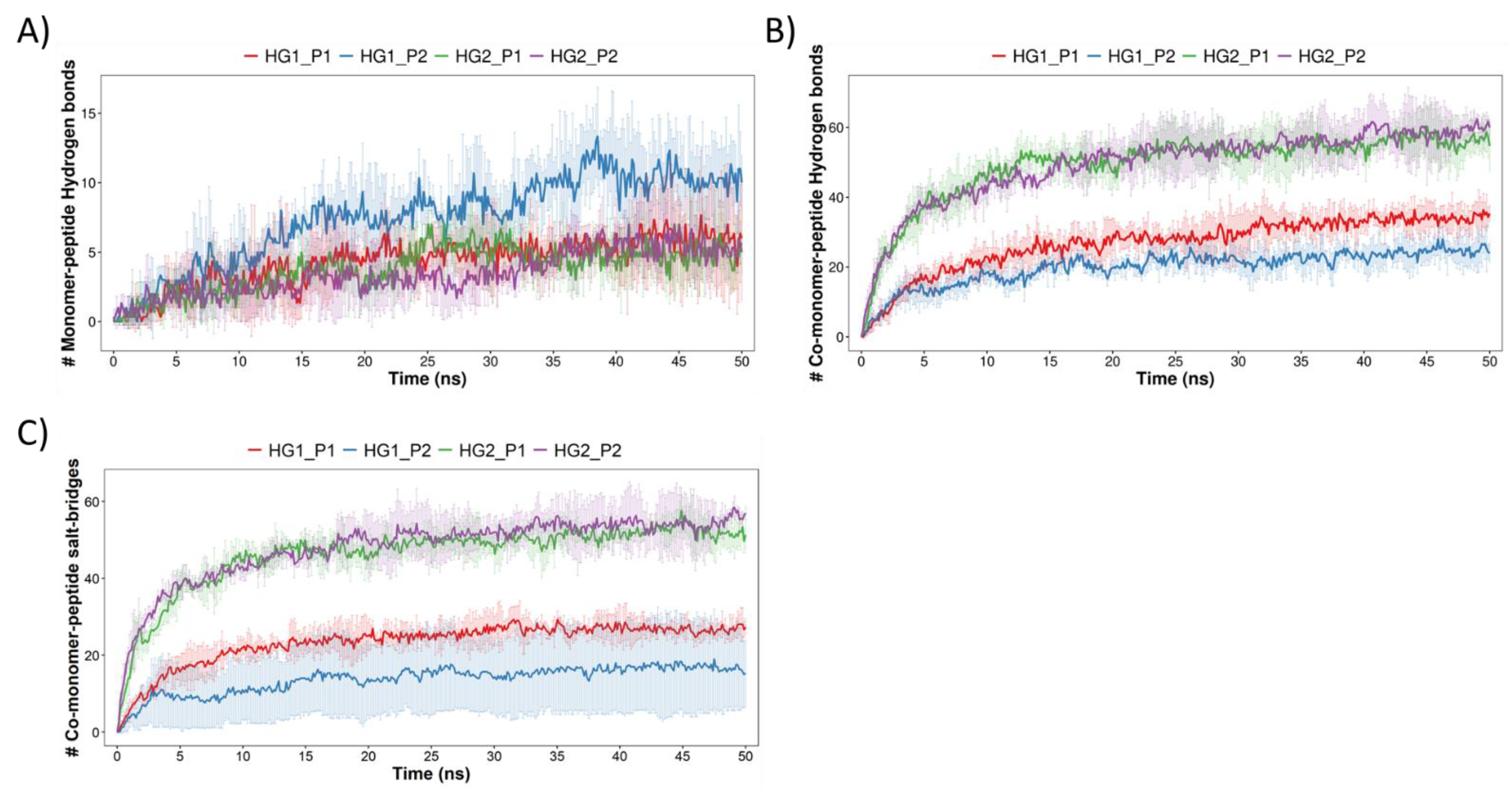
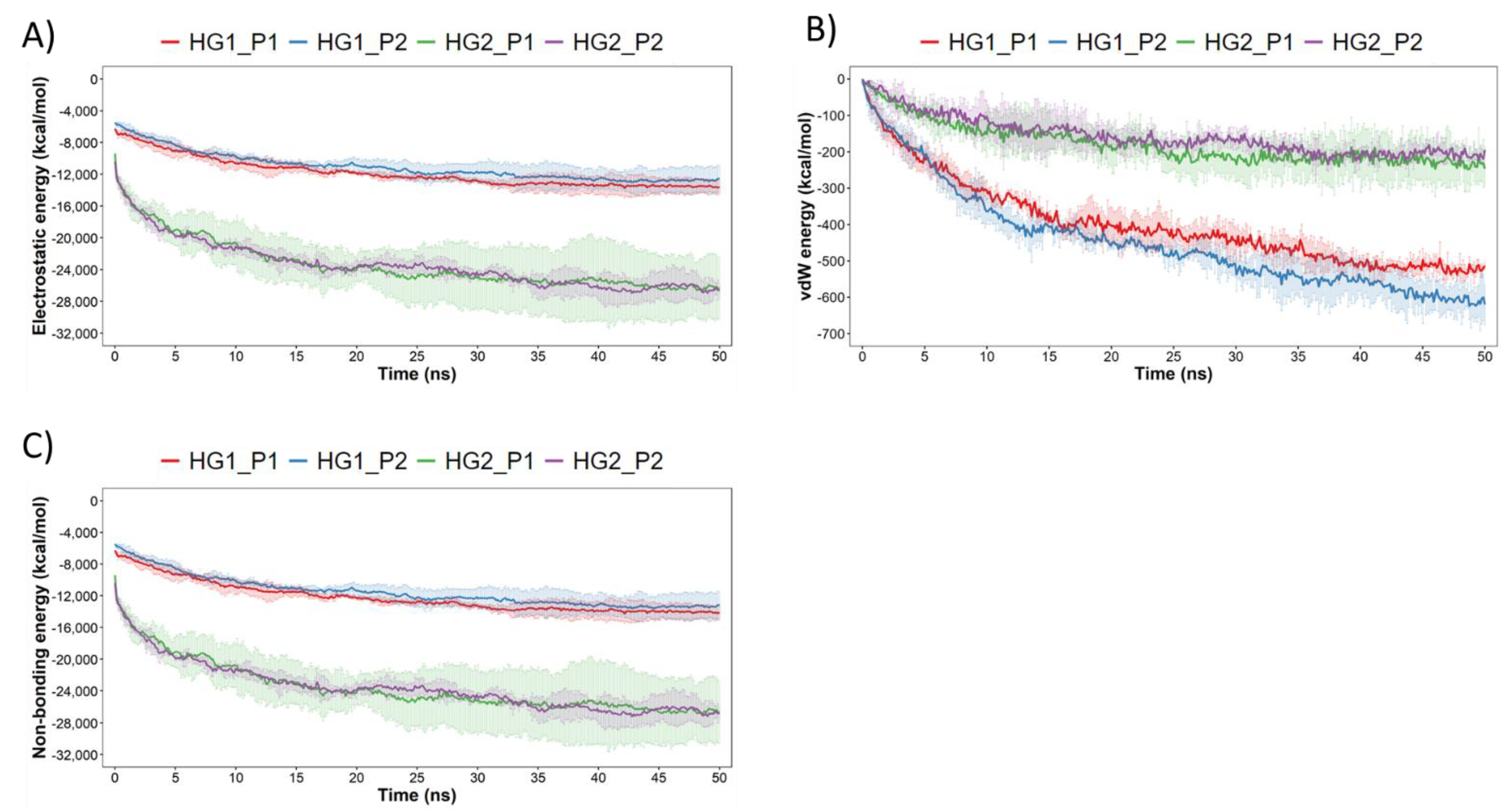
3.2. Molecular Dynamics Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knight, G.M.; Glover, R.E.; McQuaid, C.F.; Olaru, I.D.; Gallandat, K.; Leclerc, Q.J.; Fuller, N.M.; Willcocks, S.J.; Hasan, R.; van Kleef, E.; et al. Antimicrobial resistance and COVID-19: Intersections and implications. eLife 2021, 10, e64139. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; Salamat, M.K.F.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. Available online: http://www.ncbi.nlm.nih.gov/pubmed/30349322 (accessed on 19 January 2019). [CrossRef] [PubMed]
- Rice, L.B. Progress and Challenges in Implementing the Research on ESKAPE Pathogens. Infect. Control Hosp. Epidemiol. 2010, 31, S7–S10. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W.I. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci. Rep. 2018, 8, 1697. [Google Scholar] [CrossRef] [PubMed]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Hancock, R.E.; Rozek, A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 2002, 206, 143–149. [Google Scholar] [CrossRef]
- Pachón-Ibáñez, M.E.; Smani, Y.; Pachón, J.; Sánchez-Céspedes, J. Perspectives for clinical use of engineered human host defense antimicrobial peptides. FEMS Microbiol. Rev. 2017, 41, 323–342. [Google Scholar] [CrossRef]
- Martin-Serrano, Á.; Gómez, R.; Ortega, P.; De La Mata, F.J. Nanosystems as Vehicles for the Delivery of Antimicrobial Peptides (AMPs). Pharmaceutics 2019, 11, 448. [Google Scholar] [CrossRef] [PubMed]
- Borro, B.; Nordström, R.; Malmsten, M. Microgels and hydrogels as delivery systems for antimicrobial peptides. Colloids Surfaces B Biointerfaces 2020, 187, 110835. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Zhou, Y.; He, W.; Hua, D. A strategy for enhanced antibacterial activity against Staphylococcus aureus by the assembly of alamethicin with a thermo-sensitive polymeric carrier. Chem. Commun. 2016, 52, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, P.; Qi, X.; Sharif, A.R.M.; Poon, Y.F.; Cao, Y.; Chang, M.W.; Leong, S.S.J.; Chan-Park, M.B. A photopolymerized antimicrobial hydrogel coating derived from epsilon-poly-l-lysine. Biomaterials 2011, 32, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, R.; Singh, A.K.; Dhillon, A. pH-responsive drug release from dependal-M loaded polyacrylamide hydrogels. J. Sci. Adv. Mater. Devices 2017, 2, 45–50. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial Peptides and Nanotechnology, Recent Advances and Challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef]
- Pertici, V.; Pin-Barre, C.; Rivera, C.; Pellegrino, C.; Laurin, J.; Gigmes, D.; Trimaille, T. Degradable and Injectable Hydrogel for Drug Delivery in Soft Tissues. Biomacromolecules 2018, 20, 149–163. [Google Scholar] [CrossRef]
- Gao, X.; Cao, Y.; Song, X.; Zhang, Z.; Xiao, C.; He, C.; Chen, X. pH- and thermo-responsive poly(N-isopropylacrylamide-co-acrylic acid derivative) copolymers and hydrogels with LCST dependent on pH and alkyl side groups. J. Mater. Chem. B 2013, 1, 5578–5587. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Q.; Zhu, S.; Liu, H.; Chen, J. Preparation and applications of peptide-based injectable hydrogels. RSC Adv. 2019, 9, 28299–28311. [Google Scholar] [CrossRef]
- Ávila-Salas, F. An Overview of Injectable Thermo-Responsive Hydrogels and Advances in their Biomedical Applications. Curr. Med. Chem. 2020, 27, 5773–5789. [Google Scholar] [CrossRef]
- Joubert, F.; Denn, P.C.P.; Guo, Y.; Pasparakis, G. Comparison of Thermoresponsive Hydrogels Synthesized by Conventional Free Radical and RAFT Polymerization. Materials 2019, 12, 2697. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Wang, Y.; Hu, X.; Gong, H.; Li, R.; Cox, H.; Zhang, J.; Waigh, T.A.; Xu, H.; Lu, J.R. Reversible Thermoresponsive Peptide–PNIPAM Hydrogels for Controlled Drug Delivery. Biomacromolecules 2019, 20, 3601–3610. [Google Scholar] [CrossRef] [PubMed]
- Haq, M.A.; Su, Y.; Wang, D. Mechanical properties of PNIPAM based hydrogels: A review. Mater. Sci. Eng. C 2017, 70, 842–855. [Google Scholar] [CrossRef] [PubMed]
- Carreño, G.; Pereira, A.; Ávila-Salas, F.; Marican, A.; Andrade, F.; Roca-Melendres, M.M.; Valdés, O.; Vijayakumar, S.; Schwartz, S.; Abasolo, I.; et al. Development of “on-demand” thermo-responsive hydrogels for anti-cancer drugs sustained release: Rational design, in silico prediction and in vitro validation in colon cancer models. Mater. Sci. Eng. C 2021, 131, 112483. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Fasterde novostructure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-1: Maestro; Schrödinger: New York, NY, USA, 2021.
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.; Brozell, S.R.; Cerutti, D.S.; Cheatham III, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; et al. Amber 2021; University of California: San Francisco, CA, USA, 2021. [Google Scholar]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Guo, D.; Xiao, K.; Wang, X.; Wang, L.; Luo, J. A drug-specific nanocarrier design for efficient anticancer therapy. Nat. Commun. 2015, 6, 7449. [Google Scholar] [CrossRef] [PubMed]


| MP-L or P1 | NCP-3a or P2 | |
|---|---|---|
| Source | Synthetic construct | Synthetic construct |
| Family | Derived from the peptide Mastoparan | Derived from CTX-1 |
| Sequence | INLKILARLAKKIL | KLIFILSKTIPAGKNLFYKI |
| Length | 14 residues | 20 |
| Biological Activity | Anti-Gram positive, Anti-Gram negative, Antifungal | |
| Target Organism |
|
|
| Structure predicted | 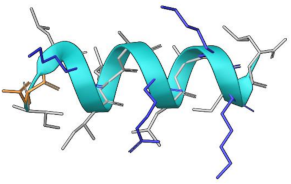 | 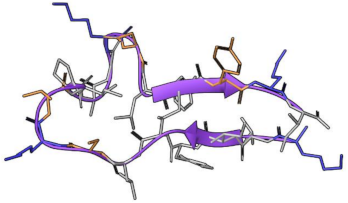 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, A.; Valdés-Muñoz, E.; Marican, A.; Cabrera-Barjas, G.; Vijayakumar, S.; Valdés, O.; Rafael, D.; Andrade, F.; Abaca, P.; Bustos, D.; et al. Rational Design of Hydrogels for Cationic Antimicrobial Peptide Delivery: A Molecular Modeling Approach. Pharmaceutics 2023, 15, 474. https://doi.org/10.3390/pharmaceutics15020474
Pereira A, Valdés-Muñoz E, Marican A, Cabrera-Barjas G, Vijayakumar S, Valdés O, Rafael D, Andrade F, Abaca P, Bustos D, et al. Rational Design of Hydrogels for Cationic Antimicrobial Peptide Delivery: A Molecular Modeling Approach. Pharmaceutics. 2023; 15(2):474. https://doi.org/10.3390/pharmaceutics15020474
Chicago/Turabian StylePereira, Alfredo, Elizabeth Valdés-Muñoz, Adolfo Marican, Gustavo Cabrera-Barjas, Sekar Vijayakumar, Oscar Valdés, Diana Rafael, Fernanda Andrade, Paulina Abaca, Daniel Bustos, and et al. 2023. "Rational Design of Hydrogels for Cationic Antimicrobial Peptide Delivery: A Molecular Modeling Approach" Pharmaceutics 15, no. 2: 474. https://doi.org/10.3390/pharmaceutics15020474
APA StylePereira, A., Valdés-Muñoz, E., Marican, A., Cabrera-Barjas, G., Vijayakumar, S., Valdés, O., Rafael, D., Andrade, F., Abaca, P., Bustos, D., & Durán-Lara, E. F. (2023). Rational Design of Hydrogels for Cationic Antimicrobial Peptide Delivery: A Molecular Modeling Approach. Pharmaceutics, 15(2), 474. https://doi.org/10.3390/pharmaceutics15020474











