Biomarkers as Biomedical Bioindicators: Approaches and Techniques for the Detection, Analysis, and Validation of Novel Biomarkers of Diseases
Abstract
:1. Introduction
2. Classification of Biomarkers
- (a)
- Prognostic biomarkers: A prognostic biomarker is used to identify the likelihood of a clinical event, disease recurrence, or disease progression in patients with a disease or medical condition.
- (b)
- Safety: A safety biomarker is measured before or after an exposure to a medical intervention or environmental agent to indicate the likelihood, presence, or extent of a pathology.
3. Biomarkers in Monitoring and Therapy of Diseases

4. Biomarker Analysis
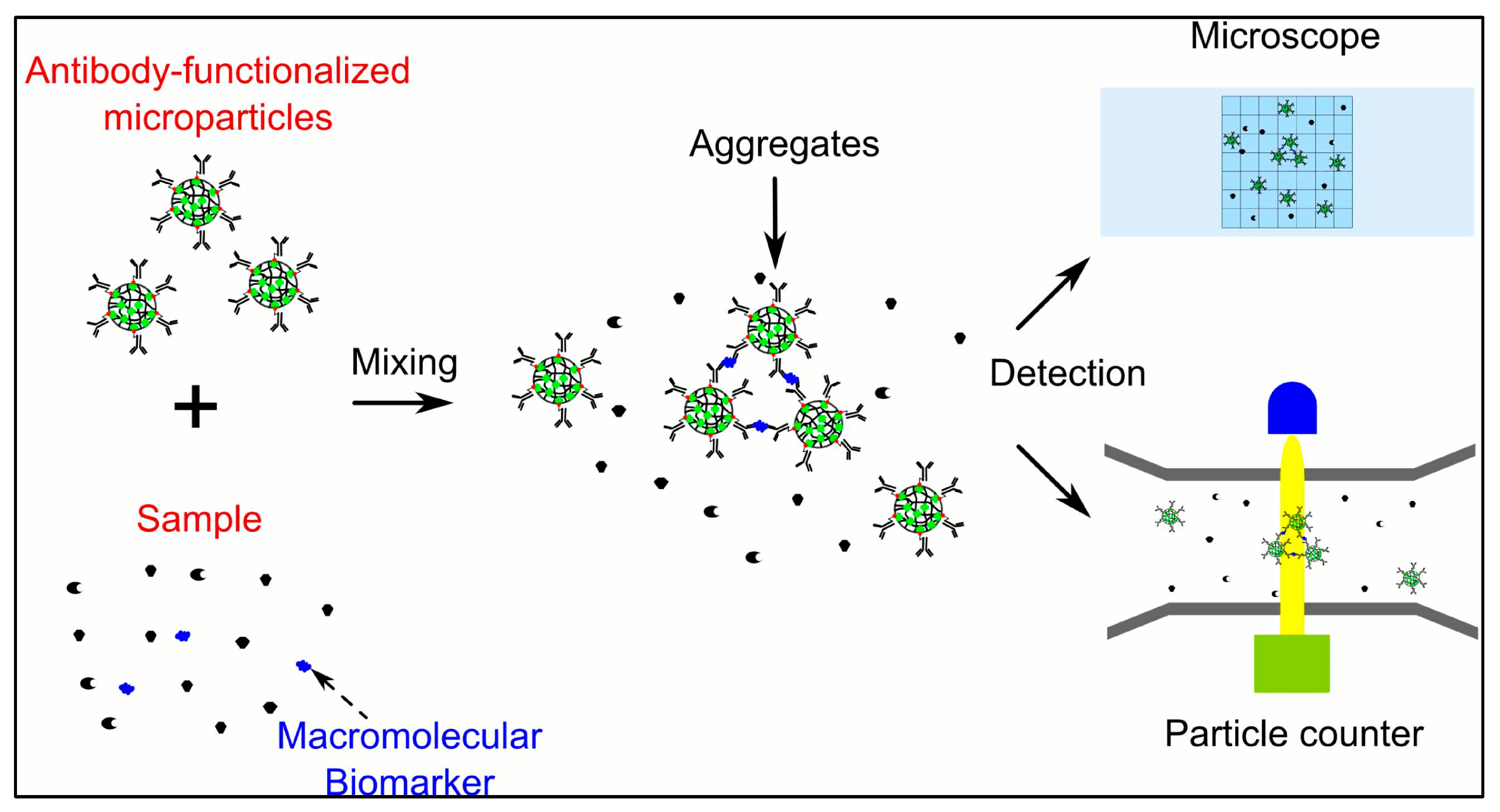
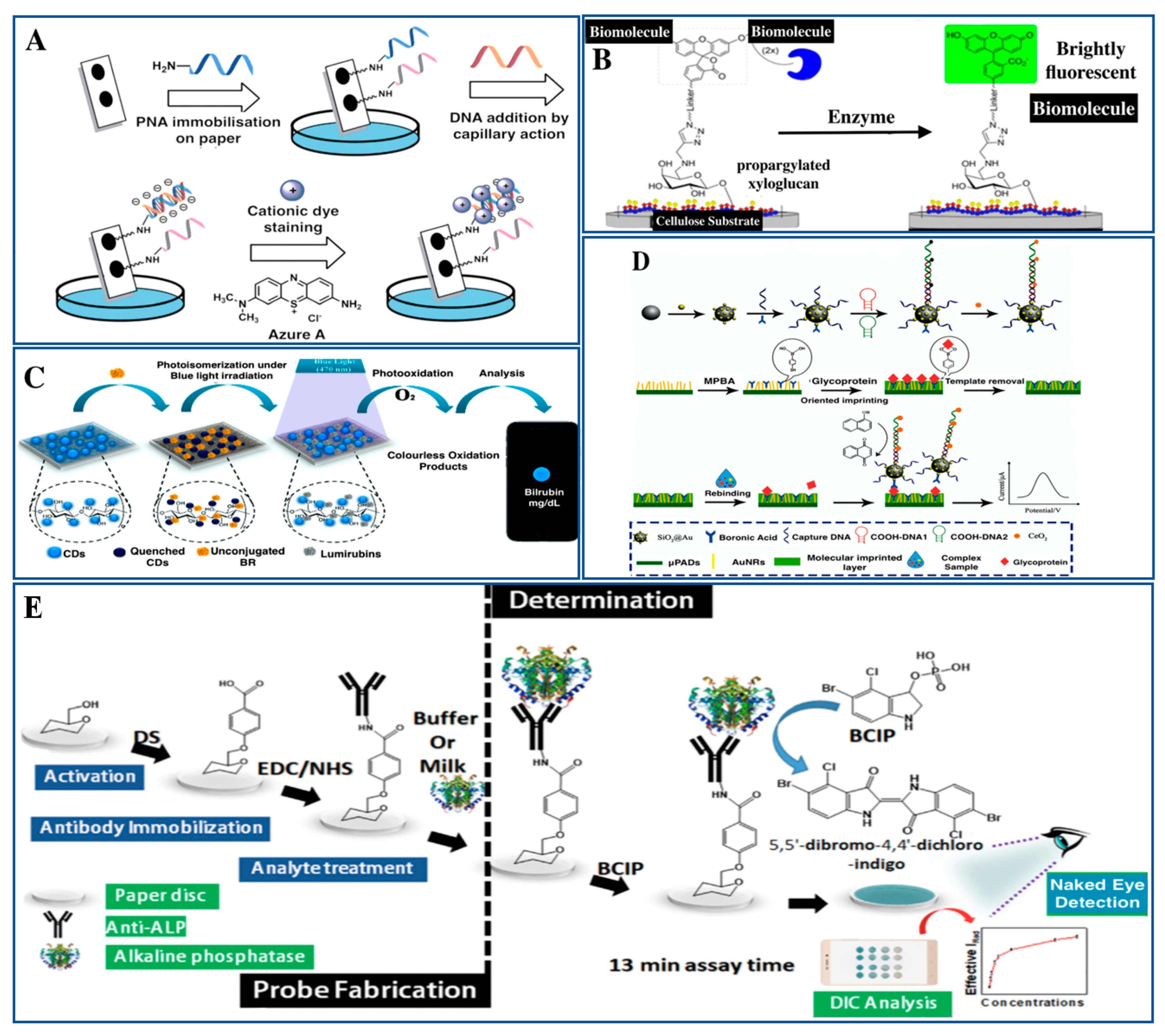
5. Approaches for Biomarkers Detection and Analysis

5.1. Surface Functionalization for Biomolecular Recognition
5.2. In Situ Analysis of Molecular Interactions
5.3. Specific Affinity Determination in Direct Binding Assays
5.4. Therapeutic Antibody Detection in Human Serum
6. Sensing of Clinically Significant Macromolecules as Potential Biomarkers
6.1. Electrochemical Biosensing of Biomolecules
6.2. Optomechanics in Biomolecular Detection
6.3. Nanotechnology in Biomolecular Sensing
7. Bioinformatics Tools and Statistical Criteria for Biomarker Analysis
8. Radiomic Markers
9. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aryutova, K.; Stoyanov, D.S.; Kandilarova, S.; Todeva-Radneva, A.; Kostianev, S.S. Clinical Use of Neurophysiological Biomarkers and Self-Assessment Scales to Predict and Monitor Treatment Response for Psychotic and Affective Disorders. Curr. Pharm. Des. 2021, 27, 4039–4048. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker Definitions and Their Applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are Biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Chen, X.-H.; Huang, S.; Kerr, D. Biomarkers in Clinical Medicine. IARC Sci. Publ. 2011, 163, 303–322. [Google Scholar]
- Ahsan, H. Biomolecules and Biomarkers in Oral Cavity: Bioassays and Immunopathology. J. Immunoass. Immunochem. 2019, 40, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Ilyin, S.E.; Belkowski, S.M.; Plata-Salamán, C.R. Biomarker Discovery and Validation: Technologies and Integrative Approaches. Trends Biotechnol. 2004, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kurian, S.; Grigoryev, Y.; Head, S.; Campbell, D.; Mondala, T.; Salomon, D.R. Applying Genomics to Organ Transplantation Medicine in Both Discovery and Validation of Biomarkers. Int. Immunopharmacol. 2007, 7, 1948–1960. [Google Scholar] [CrossRef] [PubMed]
- Silberring, J.; Ciborowski, P. Biomarker Discovery and Clinical Proteomics. TrAC Trends Anal. Chem. 2010, 29, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.D.; Verma, M.; Srivastava, S. Challenges for Biomarkers in Cancer Detection. Ann. N. Y. Acad. Sci. 2004, 1022, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary Biomarkers: Toward Future Clinical and Diagnostic Utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef]
- Mueller, W.H. Biological Markers in Epidemiology. Edited by B. S. Hulka, T.C. Wilcosky, and J. D. Griffith. xi + 236 pp. New York: Oxford University Press, 1990, $40.00 (Cloth). Am. J. Hum. Biol. 1991, 3, 218–219. [Google Scholar] [CrossRef]
- Mayeux, R. Biomarkers: Potential Uses and Limitations. Neurotherapeutics 2004, 1, 182–188. [Google Scholar] [CrossRef]
- Bhatt, A.N.; Mathur, R.; Farooque, A.; Verma, A.; Dwarakanath, B.S. Cancer Biomarkers—Current Perspectives. Indian J. Med. Res. 2010, 132, 129–149. [Google Scholar] [PubMed]
- Corella, D.; Ordovás, J.M. Biomarkers: Background, Classification and Guidelines for Applications in nutritional Epidemiology. Nutr. Hosp. 2015, 31 (Suppl. S3), 177–188. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food & Drug Administration. About Biomarkers and Qualification. 2021. Available online: https://www.fda.gov/drugs/biomarker-qualification-program/about-biomarkers-and-qualification (accessed on 20 February 2023).
- Church, R.J.; Kullak-Ublick, G.A.; Aubrecht, J.; Bonkovsky, H.L.; Chalasani, N.; Fontana, R.J.; Goepfert, J.C.; Hackman, F.; King, N.M.P.; Kirby, S.; et al. Candidate Biomarkers for the Diagnosis and Prognosis of Drug-Induced Liver Injury: An International Collaborative Effort. Hepatology 2019, 69, 760–773. [Google Scholar] [CrossRef]
- Dennis, J.K.; Sealock, J.M.; Straub, P.; Lee, Y.H.; Hucks, D.; Actkins, K.; Faucon, A.; Feng, Y.-C.A.; Ge, T.; Goleva, S.B.; et al. Clinical Laboratory Test-Wide Association Scan of Polygenic Scores Identifies Biomarkers of Complex Disease. Genome Med. 2021, 13, 6. [Google Scholar] [CrossRef]
- García-Gutiérrez, M.S.; Navarrete, F.; Sala, F.; Gasparyan, A.; Austrich-Olivares, A.; Manzanares, J. Biomarkers in Psychiatry: Concept, Definition, Types and Relevance to the Clinical Reality. Front. Psychiatry 2020, 11, 432. [Google Scholar] [CrossRef]
- Katz, R. Biomarkers and Surrogate Markers: An FDA Perspective. NeuroRx 2004, 1, 189–195. [Google Scholar] [CrossRef]
- Wickström, K.; Moseley, J. Biomarkers and Surrogate Endpoints in Drug Development: A European Regulatory View. Investig. Ophthalmol. Vis. Sci. 2017, 58, BIO27–BIO33. [Google Scholar] [CrossRef]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P.; et al. Biomarkers in Stress Related Diseases/Disorders: Diagnostic, Prognostic, and Therapeutic Values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef]
- Majkić-Singh, N. What Is a Biomarker?: From Its Discovery to Clinical Application. J. Med. Biochem. 2011, 30, 186–192. [Google Scholar] [CrossRef]
- Ray, P.; Manach, Y.L.; Riou, B.; Houle, T.T.; Warner, D.S. Statistical Evaluation of a Biomarker. Anesthesiology 2010, 112, 1023–1040. [Google Scholar] [CrossRef]
- Kumar, C.; van Gool, A.J. Chapter 1: Introduction: Biomarkers in Translational and Personalized Medicine. In Comprehensive Biomarker Discovery and Validation for Clinical Application; Royal Society of Chemistry: London, UK, 2013; pp. 3–39. [Google Scholar]
- Ensor, J.E. Biomarker Validation: Common Data Analysis Concerns. Oncologist 2014, 19, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Horvatovich, P.; Bischoff, R. Comprehensive Biomarker Discovery and Validation for Clinical Application; Royal Society of Chemistry: London, UK, 2013; ISBN 978-1-84973-422-6. [Google Scholar]
- Figg, W.D.; Newell, D.R. Pharmacological Biomarkers in the Development of Stratified Cancer Medicine. Clin. Cancer Res. 2014, 20, 2525–2529. [Google Scholar] [CrossRef] [PubMed]
- Tuntland, T.; Ethell, B.; Kosaka, T.; Blasco, F.; Zang, R.X.; Jain, M.; Gould, T.; Hoffmaster, K. Implementation of Pharmacokinetic and Pharmacodynamic Strategies in Early Research Phases of Drug Discovery and Development at Novartis Institute of Biomedical Research. Front. Pharmacol. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed]
- Huss, R. Chapter 19—Biomarkers. In Translational Regenerative Medicine; Atala, A., Allickson, J.G., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 235–241. ISBN 978-0-12-410396-2. [Google Scholar]
- Novelli, G.; Ciccacci, C.; Borgiani, P.; Papaluca Amati, M.; Abadie, E. Genetic Tests and Genomic Biomarkers: Regulation, Qualification and Validation. Clin. Cases Miner. Bone Metab. 2008, 5, 149–154. [Google Scholar] [PubMed]
- Ganesalingam, J.; Bowser, R. The Application of Biomarkers in Clinical Trials for Motor Neuron Disease. Biomark. Med. 2010, 4, 281–297. [Google Scholar] [CrossRef]
- Gosho, M.; Nagashima, K.; Sato, Y. Study Designs and Statistical Analyses for Biomarker Research. Sensors 2012, 12, 8966–8986. [Google Scholar] [CrossRef]
- Chau, C.H.; Rixe, O.; McLeod, H.; Figg, W.D. Validation of Analytical Methods for Biomarkers Employed in Drug Development. Clin. Cancer Res. 2008, 14, 5967–5976. [Google Scholar] [CrossRef]
- Gupta, R.C. Biomarkers in Toxicology; Academic Press: Boston, MA, USA, 2019. [Google Scholar]
- Ball, J.R.; Micheel, C.M. Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease; National Academies Press: Washington, DC, USA, 2010. [Google Scholar]
- Feigin, A. Evidence from Biomarkers and Surrogate Endpoints. NeuroRx 2004, 1, 323–330. [Google Scholar] [CrossRef]
- Budnik, L.T.; Adam, B.; Albin, M.; Banelli, B.; Baur, X.; Belpoggi, F.; Bolognesi, C.; Broberg, K.; Gustavsson, P.; Göen, T.; et al. Diagnosis, Monitoring and Prevention of Exposure-Related Non-Communicable Diseases in the Living and Working Environment: DiMoPEx-Project is Designed to Determine the Impacts of Environmental Exposure on Human Health. J. Occup. Med. Toxicol. 2018, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Rotman, Y.; Brown, T.A.; Hoofnagle, J.H. Evaluation of the Patient with Hepatitis B. Hepatology 2009, 49, S22–S27. [Google Scholar] [CrossRef] [PubMed]
- Group, F.-N.B.W. Monitoring Biomarker; Food and Drug Administration (US): Silver Spring, MD, USA, 2021.
- Koutsoulidou, A.; Phylactou, L.A. Circulating Biomarkers in Muscular Dystrophies: Disease and Therapy Monitoring. Mol. Ther. Methods Clin. Dev. 2020, 18, 230–239. [Google Scholar] [CrossRef]
- Tarhini, A.; Kudchadkar, R.R. Predictive and on-Treatment Monitoring Biomarkers in Advanced Melanoma: Moving toward Personalized Medicine. Cancer Treat. Rev. 2018, 71, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Shindo, Y.; Hazama, S.; Tsunedomi, R.; Suzuki, N.; Nagano, H. Novel Biomarkers for Personalized Cancer Immunotherapy. Cancers 2019, 11, 1223. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.K.; Tan, H.Y.; Saeidi, A.; Wong, W.F.; Vignesh, R.; Velu, V.; Eri, R.; Larsson, M.; Shankar, E.M. Immune Biomarkers for Diagnosis and Treatment Monitoring of Tuberculosis: Current Developments and Future Prospects. Front. Microbiol. 2019, 10, 2789. [Google Scholar] [CrossRef] [PubMed]
- Kip, A.E.; Balasegaram, M.; Beijnen, J.H.; Schellens, J.H.M.; de Vries, P.J.; Dorlo, T.P.C. Systematic Review of Biomarkers To Monitor Therapeutic Response in Leishmaniasis. Antimicrob. Agents Chemother. 2015, 59, 1–14. [Google Scholar] [CrossRef]
- Brody, T. Chapter 19—Biomarkers. In Clinical Trials, 2nd ed.; Brody, T., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 377–419. ISBN 978-0-12-804217-5. [Google Scholar]
- Oderda, G.M.; Lawless, G.D.; Wright, G.C.; Nussbaum, S.R.; Elder, R.; Kim, K.; Brixner, D.I. The Potential Impact of Monitoring Disease Activity Biomarkers on Rheumatoid Arthritis Outcomes and Costs. Pers. Med. 2018, 15, 291–301. [Google Scholar] [CrossRef]
- Giffin, R.; Robinson, S.; Olson, S. Accelerating the Development of Biomarkers for Drug Safety: Workshop Summary; National Academies Press: Washington, DC, USA, 2009. [Google Scholar]
- Lemley, K.V. An Introduction to Biomarkers: Applications to Chronic Kidney Disease. Pediatr. Nephrol. 2007, 22, 1849–1859. [Google Scholar] [CrossRef]
- Srinivas, P.R.; Kramer, B.S.; Srivastava, S. Trends in Biomarker Research for Cancer Detection. Lancet Oncol. 2001, 2, 698–704. [Google Scholar] [CrossRef]
- Ahmad, A. Customized Polymeric Nanoparticles for Targeting Inflammatory Disorders. Ph.D. Thesis, Indian Institute of Science Education and Research Mohali, Ajitgarh, India, 2021. [Google Scholar]
- Dwivedi, S.; Purohit, P.; Misra, R.; Pareek, P.; Goel, A.; Khattri, S.; Pant, K.K.; Misra, S.; Sharma, P. Diseases and Molecular Diagnostics: A Step Closer to Precision Medicine. Indian J. Clin. Biochem. 2017, 32, 374–398. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine. Improving Diagnosis in Health Care; National Academies Press: Washington, DC, USA, 2015. [Google Scholar]
- Selleck, M.J.; Senthil, M.; Wall, N.R. Making Meaningful Clinical Use of Biomarkers. Biomark. Insights 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Bensmail, H.; Haoudi, A. Postgenomics: Proteomics and Bioinformatics in Cancer Research. J. Biomed. Biotechnol. 2003, 2003, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Scholler, N.; Urban, N. CA125 in Ovarian Cancer. Biomark. Med. 2007, 1, 513–523. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Biomarkers: Hopes And Challenges in the Path From Discovery to Clinical Practice. Transl. Res. 2012, 159, 197–204. [Google Scholar] [CrossRef]
- Karley, D.; Gupta, D.; Tiwari, A. Biomarker for Cancer: A Great Promise for Future. World J. Oncol. 2011, 2, 151–157. [Google Scholar]
- Gaikwad, S.; Sawant, S.S.; Shastri, J.S. Comparison of Nonstructural Protein-1 Antigen Detection by Rapid and Enzyme-Linked Immunosorbent Assay Test and Its Correlation with Polymerase Chain Reaction for Early Diagnosis of Dengue. J. Lab. Physicians 2017, 9, 177–181. [Google Scholar] [CrossRef]
- Maxim, L.D.; Niebo, R.; Utell, M.J. Screening Tests: A Review with Examples. Inhal. Toxicol. 2014, 26, 811–828. [Google Scholar] [CrossRef]
- Al-Musalhi, K.; Al-Kindi, M.; Ramadhan, F.; Al-Rawahi, T.; Al-Hatali, K.; Mula-Abed, W.-A. Validity of Cancer Antigen-125 (CA-125) and Risk of Malignancy Index (RMI) in the Diagnosis of Ovarian Cancer. Oman Med. J. 2015, 30, 428–434. [Google Scholar] [CrossRef]
- Wang, J.; Xu, B. Targeted Therapeutic Options and Future Perspectives for HER2-Positive Breast Cancer. Signal Transduct. Target. Ther. 2019, 4, 34. [Google Scholar] [CrossRef]
- Nagpal, M.; Singh, S.; Singh, P.; Chauhan, P.; Zaidi, M.A. Tumor Markers: A Diagnostic Tool. Natl. J. Maxillofac. Surg. 2016, 7, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S. Tumor Markers in Clinical Practice: General Principles and Guidelines. Indian J. Med. Paediatr. Oncol. 2009, 30, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Al-Tarawneh, S.K.; Bencharit, S. Applications of Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight (SELDI-TOF) Mass Spectrometry in Defining Salivary Proteomic Profiles. Open Dent. J. 2009, 3, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Christians, U.; Klawitter, J.; Klepacki, J.; Klawitter, J. Chapter Four—The Role of Proteomics in the Study of Kidney Diseases and in the Development of Diagnostic Tools. In Biomarkers of Kidney Disease, 2nd ed.; Edelstein, C.L., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 119–223. ISBN 978-0-12-803014-1. [Google Scholar]
- Petricoin, E.F.; Liotta, L.A. SELDI-TOF-Based Serum Proteomic Pattern Diagnostics for Early Detection of Cancer. Curr. Opin. Biotechnol. 2004, 15, 24–30. [Google Scholar] [CrossRef]
- Simpkins, F.; Czechowicz, J.A.; Liotta, L.; Kohn, E.C. SELDI-TOF Mass Spectrometry for Cancer Biomarker Discovery and Serum Proteomic Diagnostics. Pharmacogenomics 2005, 6, 647–653. [Google Scholar] [CrossRef]
- Kosmides, A.K.; Kamisoglu, K.; Calvano, S.E.; Corbett, S.A.; Androulakis, I.P. Metabolomic Fingerprinting: Challenges and Opportunities. Crit. Rev. Biomed. Eng. 2013, 41, 205–221. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Biomarker Discovery and Translation in Metabolomics. Curr. Metab. 2013, 1, 227–240. [Google Scholar] [CrossRef]
- Hamilton, P.; O’Reilly, P.; Bankhead, P.; Abels, E.; Salto-Tellez, M. Digital and Computational Pathology for Biomarker Discovery. In Predictive Biomarkers in Oncology: Applications in Precision Medicine; Badve, S., Kumar, G.L., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 87–105. ISBN 978-3-319-95228-4. [Google Scholar]
- Upadhyay, R.K. Biomarkers in Japanese Encephalitis: A Review. Biomed. Res. Int. 2013, 2013, 591290. [Google Scholar] [CrossRef]
- Tenenbaum, J.D.; Bhuvaneshwar, K.; Gagliardi, J.P.; Hollis, K.F.; Jia, P.; Ma, L.; Nagarajan, R.; Rakesh, G.; Subbian, V.; Visweswaran, S.; et al. Translational Bioinformatics in Mental Health: Open Access Data Sources and Computational Biomarker Discovery. Brief. Bioinform. 2019, 20, 842–856. [Google Scholar] [CrossRef]
- Pinker, K.; Shitano, F.; Sala, E.; Do, R.K.; Young, R.J.; Wibmer, A.G.; Hricak, H.; Sutton, E.J.; Morris, E.A. Background, Current Role and Potential Applications of Radiogenomics. J. Magn. Reson. Imaging 2018, 47, 604–620. [Google Scholar] [CrossRef]
- Slomka, P.J.; Dey, D.; Sitek, A.; Motwani, M.; Berman, D.S.; Germano, G. Cardiac Imaging: Working towards Fully-Automated Machine Analysis & Interpretation. Expert Rev. Med. Devices 2017, 14, 197–212. [Google Scholar] [CrossRef]
- Deveci, H.S.; Kule, M.; Kule, Z.A.; Habesoglu, T.E. Diagnostic Challenges in Cervical Tuberculous Lymphadenitis: A Review. North. Clin. Istanb. 2016, 3, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Gharekhanloo, F.; Haseli, M.M.; Torabian, S. Value of Ultrasound in the Detection of Benign and Malignant Breast Diseases: A Diagnostic Accuracy Study. Oman Med. J. 2018, 33, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Mesri, M. Advances in Proteomic Technologies and Its Contribution to the Field of Cancer. Adv. Med. 2014, 2014, 238045. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of Bioactive Metabolites Using Activity Metabolomics. Nat. Rev. Mol. Cell. Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, N. In Vivo Magnetic Resonance Techniques and Drug Discovery. Braz. J. Phys. 2006, 36, 16–22. [Google Scholar] [CrossRef]
- Seyhan, A.A. Biomarkers in Drug Discovery and Development. Eur. Pharm. Rev. 2010, 5, 19–25. [Google Scholar]
- Zhang, X.; Li, Q.; Xu, Z.; Dou, J. Mass Spectrometry-Based Metabolomics in Health and Medical Science: A Systematic Review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef]
- Scaros, O.; Fisler, R. Biomarker Technology Roundup: From Discovery to Clinical Applications, a Broad Set of Tools is Required to Translate from the Lab to the Clinic. BioTechniques 2005, 38, S30–S32. [Google Scholar] [CrossRef]
- Wu, H.; Han, Y.; Yang, X.; Chase, G.G.; Tang, Q.; Lee, C.-J.; Cao, B.; Zhe, J.; Cheng, G. A Versatile Microparticle-Based Immunoaggregation Assay for Macromolecular Biomarker Detection and Quantification. PLoS ONE 2015, 10, e0115046. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer Biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of Exosomal Proteins in Cancer Diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Conrads, T.P.; Veenstra, T.D. Proteomics Approaches to Biomarker Detection. Brief. Funct. Genom. 2005, 4, 69–75. [Google Scholar] [CrossRef]
- Ray, S.; Reddy, P.J.; Choudhary, S.; Raghu, D.; Srivastava, S. Emerging Nanoproteomics Approaches for Disease Biomarker Detection: A Current Perspective. J. Proteom. 2011, 74, 2660–2681. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Imran, M.; Sharma, N. Precision Nanotoxicology in Drug Development: Current Trends and Challenges in Safety and Toxicity Implications of Customized Multifunctional Nanocarriers for Drug-Delivery Applications. Pharmaceutics 2022, 14, 2463. [Google Scholar] [CrossRef]
- Ahmad, A. Safety and Toxicity Implications of Multifunctional Drug Delivery Nanocarriers on Reproductive Systems In Vitro and In Vivo. Front. Toxicol. 2022, 4, 895667. [Google Scholar] [CrossRef]
- Skelley, A.M.; Scherer, J.R.; Aubrey, A.D.; Grover, W.H.; Ivester, R.H.C.; Ehrenfreund, P.; Grunthaner, F.J.; Bada, J.L.; Mathies, R.A. Development and Evaluation of a Microdevice for Amino Acid Biomarker Detection and Analysis on Mars. PNAS 2005, 102, 1041–1046. [Google Scholar] [CrossRef]
- Rai, V.; Mukherjee, R.; Ghosh, A.K.; Routray, A.; Chakraborty, C. “Omics” in Oral Cancer: New Approaches for Biomarker Discovery. Arch. Oral Biol. 2018, 87, 15–34. [Google Scholar] [CrossRef]
- Adam, B.-L.; Vlahou, A.; Semmes, O.J.; Wright, G.L., Jr. Proteomic Approaches to Biomarker Discovery in Prostate and Bladder Cancers. Proteomics 2001, 1, 1264–1270. [Google Scholar] [CrossRef]
- Maruvada, P.; Wang, W.; Wagner, P.D.; Srivastava, S. Biomarkers in Molecular Medicine: Cancer Detection and Diagnosis. BioTechniques 2005, 38, S9–S15. [Google Scholar] [CrossRef]
- Bezinge, L.; Suea-Ngam, A.; de Mello, A.J.; Shih, C.-J. Nanomaterials for Molecular Signal Amplification in Electrochemical Nucleic acid Biosensing: Recent Advances and Future Prospects for Point-of-Care Diagnostics. Mol. Syst. Des. Eng. 2020, 5, 49–66. [Google Scholar] [CrossRef]
- Kocakulak, N.A.; Saygin, A.S. The Importance of Nano Biosensors and Ethical Elements in Sports Performance Analysis. Nat. Appl. Sci. J. 2020, 3, 17–27. [Google Scholar]
- Nagamune, T. Biomolecular Engineering for Nanobio/Bionanotechnology. Nano Converg. 2017, 4, 9. [Google Scholar] [CrossRef]
- Spicer, C.D.; Pashuck, E.T.; Stevens, M.M. Achieving Controlled Biomolecule–Biomaterial Conjugation. Chem. Rev. 2018, 118, 7702–7743. [Google Scholar] [CrossRef]
- Morales, M.A.; Mark Halpern, J. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef]
- Hnaien, M.; Diouani, M.F.; Helali, S.; Hafaid, I.; Hassen, W.M.; Renault, N.J.; Ghram, A.; Abdelghani, A. Immobilization of Specific Antibody on SAM Functionalized Gold Electrode for Rabies Virus Detection by Electrochemical Impedance Spectroscopy. Biochem. Eng. J. 2008, 39, 443–449. [Google Scholar] [CrossRef]
- Nangare, S.N.; Patil, P.O. Affinity-Based Nanoarchitectured Biotransducer for Sensitivity Enhancement of Surface Plasmon Resonance Sensors for In Vitro Diagnosis: A Review. ACS Biomater. Sci. Eng. 2020, 7, 2–30. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Yasui, T.; Yokoyama, A.; Goda, T.; Hara, M.; Yanagida, T.; Kaji, N.; Kanai, M.; Nagashima, K.; Miyahara, Y. Biomolecular Recognition on Nanowire Surfaces Modified by the Self-Assembled Monolayer. Lab A Chip 2018, 18, 3225–3229. [Google Scholar] [CrossRef] [PubMed]
- Oliverio, M.; Perotto, S.; Messina, G.C.; Lovato, L.; De Angelis, F. Chemical Functionalization of Plasmonic Surface Biosensors: A Tutorial Review on Issues, Strategies, and Costs. ACS Appl. Mater. Interfaces 2017, 9, 29394–29411. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, M.D.; Nimse, S.B. Surface Modification Chemistries of Materials Used in Diagnostic Platforms with Biomolecules. J. Chem. 2016, 2016, 9241378. [Google Scholar] [CrossRef]
- Almeida, A.; Rosa, A.M.M.; Azevedo, A.M.; Prazeres, D.M.F. A Biomolecular Recognition Approach for the Functionalization of Cellulose with Gold Nanoparticles. J. Mol. Recognit. 2017, 30, e2634. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.E.; Tse, K.-Y.; Hindin, E.; Nichols, B.M.; Lasseter Clare, T.; Hamers, R.J. Covalent Functionalization for Biomolecular Recognition on Vertically Aligned Carbon Nanofibers. Chem. Mater. 2005, 17, 4971–4978. [Google Scholar] [CrossRef]
- Tavallaie, R.; McCarroll, J.; Le Grand, M.; Ariotti, N.; Schuhmann, W.; Bakker, E.; Tilley, R.D.; Hibbert, D.B.; Kavallaris, M.; Gooding, J.J. Nucleic Acid Hybridization on an Electrically Reconfigurable Network of Gold-Coated Magnetic Nanoparticles Enables MicroRNA Detection in Blood. Nat. Nanotechnol. 2018, 13, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Hausdorf, D.; Schalper, K.A.; Neumeister, V.; Rimm, D.L. Quantitative Measurement of Cancer Tissue Biomarkers in the Lab and in the Clinic. Lab. Investig. 2015, 95, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Gatterdam, V.; Frutiger, A.; Stengele, K.-P.; Heindl, D.; Lübbers, T.; Vörös, J.; Fattinger, C. Focal Molography is a New Method for the In Situ Analysis of Molecular Interactions in Biological Samples. Nat. Nanotechnol. 2017, 12, 1089–1095. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Le Clair, S.V.; Ye, S.; Chen, Z. Molecular Interactions between Magainin 2 and Model Membranes in Situ. J. Phys. Chem. B 2009, 113, 12358–12363. [Google Scholar] [CrossRef]
- Künneke, S.; Janshoff, A. Visualization of Molecular Recognition Events on Microstructured Lipid-Membrane Compartments by In Situ Scanning Force Microscopy. Angew. Chem. Int. Ed. 2002, 41, 314–316. [Google Scholar] [CrossRef]
- Ivnitski, D.; Wolf, T.; Solomon, B.; Fleminger, G.; Rishpon, J. An Amperometric Biosensor for Real-Time Analysis of Molecular Recognition. Bioelectrochemistry Bioenerg. 1998, 45, 27–32. [Google Scholar] [CrossRef]
- Manolova, M.; Boyen, H.-G.; Kucera, J.; Groß, A.; Romanyuk, A.; Oelhafen, P.; Ivanova, V.; Kolb, D.M. Chemical Interactions at Metal/Molecule Interfaces in Molecular Junctions—A Pathway Towards Molecular Recognition. Adv. Mater. 2009, 21, 320–324. [Google Scholar] [CrossRef]
- Ahmad, A.; Swami, R.; Sharma, T.; Jain, A. Nanotechnology in the Management of Hormonal Cancer. In Hormone Related Cancer Mechanistic and Nanomedicines: Challenges and Prospects; Rahman, M.H., Almalki, W., Alrobaian, M., Beg, S., Alharbi, K.S., Eds.; Springer Nature: Singapore, 2022; pp. 13–48. ISBN 978-981-19555-8-7. [Google Scholar]
- Cazares, L.H.; Troyer, D.A.; Wang, B.; Drake, R.R.; Semmes, O.J. MALDI Tissue Imaging: From Biomarker Discovery to Clinical Applications. Anal. Bioanal. Chem. 2011, 401, 17–27. [Google Scholar] [CrossRef]
- Ahmad, A.; Bulani, Y.; Sharma, S.S. Naringenin Shows Ameliorative Effects in Isoproterenol-induced Myocardial Infarction. In Proceedings of the 12th Annual Conference of International Society of Heart Research (ISHRCON), New Delhi, India, 14–15 March 2015. [Google Scholar]
- Jia, M.; Li, S.; Zang, L.; Lu, X.; Zhang, H. Analysis of Biomolecules Based on the Surface Enhanced Raman Spectroscopy. Nanomaterials 2018, 8, 730. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 Infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, S.; Xu, T.; Bodenko, V.; Orlova, A.; Oroujeni, M.; Rinne, S.S.; Tolmachev, V.; Vorobyeva, A.; Gräslund, T. Preclinical Evaluation of a New Format of 68Ga-and 111In-Labeled Affibody Molecule ZIGF-1R: 4551 for the Visualization of IGF-1R Expression in Malignant Tumors Using PET and SPECT. Pharmaceutics 2022, 14, 1475. [Google Scholar] [CrossRef]
- Komane, P.P.; Kumar, P.; Choonara, Y.E. Atrial Natriuretic Peptide Antibody-Functionalised, PEGylated Multiwalled Carbon Nanotubes for Targeted Ischemic Stroke Intervention. Pharmaceutics 2021, 13, 1357. [Google Scholar] [CrossRef]
- Beeg, M.; Nobili, A.; Orsini, B.; Rogai, F.; Gilardi, D.; Fiorino, G.; Danese, S.; Salmona, M.; Garattini, S.; Gobbi, M. A Surface Plasmon Resonance-Based Assay to Measure Serum Concentrations of Therapeutic Antibodies and Anti-Drug Antibodies. Sci. Rep. 2019, 9, 2064. [Google Scholar] [CrossRef] [PubMed]
- Stubenrauch, K.; Wessels, U.; Lenz, H. Evaluation of an Immunoassay for Human-Specific Quantitation of Therapeutic Antibodies in Serum Samples from Non-Human Primates. J. Pharm. Biomed. Anal. 2009, 49, 1003–1008. [Google Scholar] [CrossRef]
- Fischer, S.K.; Yang, J.; Anand, B.; Cowan, K.; Hendricks, R.; Li, J.; Nakamura, G.; Song, A. The Assay Design Used for Measurement of Therapeutic Antibody Concentrations Can Affect Pharmacokinetic Parameters. MAbs 2012, 4, 623–631. [Google Scholar] [CrossRef]
- Divya; Mahapatra, S.; Srivastava, V.R.; Chandra, P. Nanobioengineered Sensing Technologies Based on Cellulose Matrices for Detection of Small Molecules, Macromolecules, and Cells. Biosensors 2021, 11, 168. [Google Scholar] [CrossRef]
- Barr, A.J. The Biochemical Basis of Disease. Essays Biochem. 2018, 62, 619–642. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef] [PubMed]
- Barry, R.C.; Lin, Y.; Wang, J.; Liu, G.; Timchalk, C.A. Nanotechnology-Based Electrochemical Sensors for Biomonitoring Chemical Exposures. J. Expo. Sci. Environ. Epidemiol. 2009, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Jackowska, K.; Krysinski, P. New Trends in the Electrochemical Sensing of Dopamine. Anal. Bioanal. Chem. 2013, 405, 3753–3771. [Google Scholar] [CrossRef]
- Malhotra, B.D.; Ali, M.A. Nanomaterials in Biosensors. In Nanomaterials for Biosensors; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1–74. [Google Scholar] [CrossRef]
- Li, T.; Shang, D.; Gao, S.; Wang, B.; Kong, H.; Yang, G.; Shu, W.; Xu, P.; Wei, G. Two-Dimensional Material-Based Electrochemical Sensors/Biosensors for Food Safety and Biomolecular Detection. Biosensors 2022, 12, 314. [Google Scholar] [CrossRef] [PubMed]
- Suhito, I.R.; Koo, K.-M.; Kim, T.-H. Recent Advances in Electrochemical Sensors for the Detection of Biomolecules and Whole Cells. Biomedicines 2020, 9, 15. [Google Scholar] [CrossRef]
- Tabish, T.A.; Hayat, H.; Abbas, A.; Narayan, R.J. Graphene Quantum Dot–Based Electrochemical Biosensing for Early Cancer Detection. Curr. Opin. Electrochem. 2021, 30, 100786. [Google Scholar] [CrossRef]
- Kour, R.; Arya, S.; Young, S.-J.; Gupta, V.; Bandhoria, P.; Khosla, A. Recent Advances in Carbon Nanomaterials as Electrochemical Biosensors. J. Electrochem. Soc. 2020, 167, 037555. [Google Scholar] [CrossRef]
- Adams, A.; Malkoc, A.; La Belle, J.T. The Development of a Glucose Dehydrogenase 3D-Printed Glucose Sensor: A Proof-of-Concept Study. J. Diabetes Sci. Technol. 2017, 12, 176–182. [Google Scholar] [CrossRef]
- Otero, F.; Magner, E. Biosensors—Recent Advances and Future Challenges in Electrode Materials. Sensors 2020, 20, 3561. [Google Scholar] [CrossRef]
- Liu, B.; Ouyang, X.; Ding, Y.; Luo, L.; Xu, D.; Ning, Y. Electrochemical Preparation of Nickel and Copper Oxides-Decorated Graphene Composite for Simultaneous Determination of Dopamine, Acetaminophen and Tryptophan. Talanta 2016, 146, 114–121. [Google Scholar] [CrossRef]
- Han, T.; Kundu, S.; Nag, A.; Xu, Y. 3D Printed Sensors for Biomedical Applications: A Review. Sensors 2019, 19, 1706. [Google Scholar] [CrossRef]
- Asif, M.; Aziz, A.; Azeem, M.; Wang, Z.; Ashraf, G.; Xiao, F.; Chen, X.; Liu, H. A Review on Electrochemical Biosensing Platform Based on Layered Double Hydroxides for Small Molecule Biomarkers Determination. Adv. Colloid. Interface Sci. 2018, 262, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, W.; Ren, Z.; Ma, Y.; Dong, B.; Zhou, G.; Lee, C. Progress of Optomechanical Micro/Nano Sensors: A Review. Int. J. Optomechatronics 2021, 15, 120–159. [Google Scholar] [CrossRef]
- Li, T.; Wang, W.; Yi, X. Enhancing the Sensitivity of Optomechanical Mass Sensors with a Laser in a Squeezed State. Phys. Rev. A 2021, 104, 013521. [Google Scholar] [CrossRef]
- Vollmer, F.; Yang, L. Label-Free Detection with High-Q Microcavities: A Review of Biosensing Mechanisms for Integrated Devices. Nanophotonics 2012, 1, 267–291. [Google Scholar] [CrossRef]
- Forstner, S.; Knittel, J.; Sheridan, E.; Swaim, J.D.; Rubinsztein-Dunlop, H.; Bowen, W.P. Sensitivity and Performance of Cavity Optomechanical Field Sensors. Photonic Sens. 2012, 2, 259–270. [Google Scholar] [CrossRef]
- Yu, W.; Jiang, W.C.; Lin, Q.; Lu, T. Cavity Optomechanical Spring Sensing of Single Molecules. Nat. Commun. 2016, 7, 12311. [Google Scholar] [CrossRef]
- Kosaka, P.M.; Calleja, M.; Tamayo, J. Optomechanical Devices for Deep Plasma Cancer Proteomics. Semin. Cancer Biol. 2018, 52, 26–38. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating Tumor DNA: A Promising Biomarker in the Liquid Biopsy of Cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Kobun, R. (Ed.) Chapter 1—Biosensor and Nanotechnology. In Advanced Food Analysis Tools; Academic Press: Boston, MA, USA, 2021; pp. 1–18. ISBN 978-0-12-820591-4. [Google Scholar]
- Jianrong, C.; Yuqing, M.; Nongyue, H.; Xiaohua, W.; Sijiao, L. Nanotechnology and Biosensors. Biotechnol. Adv. 2004, 22, 505–518. [Google Scholar] [CrossRef]
- Hasan, A.; Nurunnabi, M.; Morshed, M.; Paul, A.; Polini, A.; Kuila, T.; Al Hariri, M.; Lee, Y.; Jaffa, A.A. Recent Advances in Application of Biosensors in Tissue Engineering. Biomed. Res. Int. 2014, 2014, 307519. [Google Scholar] [CrossRef]
- Shetti, N.P.; Mishra, A.; Bukkitgar, S.D.; Basu, S.; Narang, J.; Raghava Reddy, K.; Aminabhavi, T.M. Conventional and Nanotechnology-Based Sensing Methods for SARS Coronavirus (2019-NCoV). ACS Appl. Bio Mater. 2021, 4, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, A.; Nine, M.J.; Silva, F.S. Biosensing Platform on Ferrite Magnetic Nanoparticles: Synthesis, Functionalization, Mechanism and Applications. Adv. Colloid Interface Sci. 2021, 290, 102380. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ye, X.; Cui, T. Recent Progress of Biomarker Detection Sensors. Research 2020, 2020, 7949037. [Google Scholar] [CrossRef] [PubMed]
- Sharifianjazi, F.; Rad, A.J.; Bakhtiari, A.; Niazvand, F.; Esmaeilkhanian, A.; Bazli, L.; Abniki, M.; Irani, M.; Moghanian, A. Biosensors and Nanotechnology for Cancer Diagnosis (Lung and Bronchus, Breast, Prostate, and Colon): A Systematic Review. Biomed. Mater. 2021, 17, 012002. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gui, Y.; Lv, X.; He, J.; Xie, F.; Li, J.; Cai, J. Nanomaterial-Based Fluorescent Biosensor for Food Safety Analysis. Biosensors 2022, 12, 1072. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Güntherodt, H.-J.; Gerber, C.; Gimzewski, J.K. Translating Biomolecular Recognition into Nanomechanics. Science 2000, 288, 316–318. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Lorentzen, M.; Kjems, J.; Besenbacher, F. Nanomechanical Sensing of DNA Sequences Using Piezoresistive Cantilevers. Langmuir 2005, 21, 8400–8408. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular Uptake of Nanoparticles: Journey Inside the Cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, G.; Zhang, L.; Wang, Q.; Li, H.; Han, Y.; Xie, L.; Yan, Z.; Li, Y.; An, Y.; et al. Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis. Front. Oncol. 2020, 10, 68. [Google Scholar] [CrossRef]
- Zappia, L.; Phipson, B.; Oshlack, A. Exploring the Single-Cell RNA-Seq Analysis Landscape with the scRNA-Tools Database. PLoS Comput. Biol. 2018, 14, e1006245. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Peng, M.; Tang, L.; Ouyang, J.; Xiong, F.; Guo, C.; Tang, Y.; Zhou, Y.; Liao, Q.; et al. Single-Cell RNA Sequencing in Cancer Research. J. Exp. Clin. Cancer Res. 2021, 40, 81. [Google Scholar] [CrossRef]
- Chevalier, A.; Yang, S.; Khurshid, Z.; Sahelijo, N.; Tong, T.; Huggins, J.H.; Yajima, M.; Campbell, J.D. The Mutational Signature Comprehensive Analysis Toolkit (Musicatk) for the Discovery, Prediction, and Exploration of Mutational Signatures. Cancer Res. 2021, 81, 5813–5817. [Google Scholar] [CrossRef] [PubMed]
- Maura, F.; Degasperi, A.; Nadeu, F.; Leongamornlert, D.; Davies, H.; Moore, L.; Royo, R.; Ziccheddu, B.; Puente, X.S.; Avet-Loiseau, H.; et al. A Practical Guide for Mutational Signature Analysis in Hematological Malignancies. Nat. Commun. 2019, 10, 2969. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, R.T.; Mattiolo, P.; Mafficini, A.; Hong, S.-M.; Piredda, M.L.; Taormina, S.V.; Malleo, G.; Marchegiani, G.; Pea, A.; Salvia, R.; et al. Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Pancreatic Cancer: Systematic Review and Still-Open Questions. Cancers 2021, 13, 3119. [Google Scholar] [CrossRef]
- Al-Harazi, O.; Kaya, I.H.; El Allali, A.; Colak, D. A Network-Based Methodology to Identify Subnetwork Markers for Diagnosis and Prognosis of Colorectal Cancer. Front. Genet. 2021, 12, 721949. [Google Scholar] [CrossRef]
- Hofree, M.; Shen, J.P.; Carter, H.; Gross, A.; Ideker, T. Network-Based Stratification of Tumor Mutations. Nat. Methods 2013, 10, 1108–1115. [Google Scholar] [CrossRef]
- Koh, D.-M.; Papanikolaou, N.; Bick, U.; Illing, R.; Kahn, C.E.; Kalpathi-Cramer, J.; Matos, C.; Martí-Bonmatí, L.; Miles, A.; Mun, S.K.; et al. Artificial Intelligence and Machine Learning in Cancer Imaging. Commun. Med. 2022, 2, 133. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine Learning Applications in Cancer Prognosis and Prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Daza, J.; Itzel, T.; Betge, J.; Zhan, T.; Marmé, F.; Teufel, A. Prognostic Cancer Gene Expression Signatures: Current Status and Challenges. Cells 2021, 10, 648. [Google Scholar] [CrossRef] [PubMed]
- Buzdin, A.; Tkachev, V.; Zolotovskaia, M.; Garazha, A.; Moshkovskii, S.; Borisov, N.; Gaifullin, N.; Sorokin, M.; Suntsova, M. Chapter One—Using Proteomic and Transcriptomic Data to Assess Activation of Intracellular Molecular Pathways. In Advances in Protein Chemistry and Structural Biology; Donev, R., Karabencheva-Christova, T., Eds.; Proteomics and Systems Biology; Academic Press: Boston, MA, USA, 2021; Volume 127, pp. 1–53. [Google Scholar]
- Zolotovskaia, M.A.; Kovalenko, M.A.; Tkachev, V.S.; Simonov, A.M.; Sorokin, M.I.; Kim, E.; Kuzmin, D.V.; Karademir-Yilmaz, B.; Buzdin, A.A. Next-Generation Grade and Survival Expression Biomarkers of Human Gliomas Based on Algorithmically Reconstructed Molecular Pathways. Int. J. Mol. Sci. 2022, 23, 7330. [Google Scholar] [CrossRef] [PubMed]
- Ou, F.-S.; Michiels, S.; Shyr, Y.; Adjei, A.A.; Oberg, A.L. Biomarker Discovery and Validation: Statistical Considerations. J. Thorac. Oncol. 2021, 16, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Fornacon-Wood, I.; Faivre-Finn, C.; O’Connor, J.P.B.; Price, G.J. Radiomics As a Personalized Medicine Tool in Lung Cancer: Separating the Hope from the Hype. Lung Cancer 2020, 146, 197–208. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Z.; He, L.; Chen, X.; Pan, D.; Ma, Z.; Liang, C.; Tian, J.; Liang, C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non—Small Cell Lung Cancer. Radiology 2016, 281, 947–957. [Google Scholar] [CrossRef]
- Bogowicz, M.; Leijenaar, R.T.H.; Tanadini-Lang, S.; Riesterer, O.; Pruschy, M.; Studer, G.; Unkelbach, J.; Guckenberger, M.; Konukoglu, E.; Lambin, P. Post-Radiochemotherapy PET Radiomics in Head and Neck Cancer—The Influence of Radiomics Implementation on the Reproducibility of Local Control Tumor Models. Radiother. Oncol. 2017, 125, 385–391. [Google Scholar] [CrossRef]
- Bogowicz, M.; Riesterer, O.; Ikenberg, K.; Stieb, S.; Moch, H.; Studer, G.; Guckenberger, M.; Tanadini-Lang, S. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 921–928. [Google Scholar] [CrossRef]
- Fan, M.; Li, H.; Wang, S.; Zheng, B.; Zhang, J.; Li, L. Radiomic Analysis Reveals DCE-MRI Features for Prediction of Molecular Subtypes of Breast Cancer. PLoS ONE 2017, 12, e0171683. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wu, M.; Ye, Z. Imaging-Based Biomarkers for Predicting and Evaluating Cancer Immunotherapy Response. Radiol. Imaging Cancer 2019, 1, e190031. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, Y.; Zha, X.; Bao, J.; Wu, Q.; Dai, H.; Hu, C. A Combination of Radiomic Features, Imaging Characteristics, and Serum Tumor Biomarkers to Predict the Possibility of the High-Grade Subtypes of Lung Adenocarcinoma. Acad. Radiol. 2022, 29, 1792–1801. [Google Scholar] [CrossRef] [PubMed]



| Biomarkers | Classification |
|---|---|
| Epigenetic | Biomarkers based on epigenetics, e.g., DNA methylation, histone modification, non-coding RNAs |
| Genetic | Biomarkers based on changes in DNA, e.g., polymorphism of a single nucleotide (SNP) |
| Lipidomic | Biomarkers based on the lipid profile |
| Metabolomic | Biomarkers based on the metabolic profile |
| Proteomic | Biomarkers based on the protein profile |
| Transcriptomic | Biomarkers based on RNA profile, e.g., expression of RNA |
| Metrics | Details |
|---|---|
| Sensitivity | The proportion of cases that test positive |
| Specificity | The proportion of controls that test negative |
| Positive predictive value | Proportion of test-positive patients who actually have the disease; is a function of disease prevalence |
| Negative predictive value | Proportion of test-negative patients who truly do not have the disease; is a function of disease prevalence |
| ROC (plot of sensitivity (true-positive rate) versus 1–specificity (false-positive rate), with a data point calculated for every value of the marker in the data set) curve | Plot of sensitivity (true-positive rate) versus 1–specificity (false-positive rate), with a data point calculated for every value of the marker in the data set |
| Discrimination | How well the marker distinguishes cases from controls; often measured by the area under the ROC curve; ranges from 0 to 1, with 0.5 indicating performance equivalent to a coin flip and 1 corresponding to perfect ability to distinguish |
| Calibration | How well a marker estimates the risk of disease or of the event of interest |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, A.; Imran, M.; Ahsan, H. Biomarkers as Biomedical Bioindicators: Approaches and Techniques for the Detection, Analysis, and Validation of Novel Biomarkers of Diseases. Pharmaceutics 2023, 15, 1630. https://doi.org/10.3390/pharmaceutics15061630
Ahmad A, Imran M, Ahsan H. Biomarkers as Biomedical Bioindicators: Approaches and Techniques for the Detection, Analysis, and Validation of Novel Biomarkers of Diseases. Pharmaceutics. 2023; 15(6):1630. https://doi.org/10.3390/pharmaceutics15061630
Chicago/Turabian StyleAhmad, Anas, Mohammad Imran, and Haseeb Ahsan. 2023. "Biomarkers as Biomedical Bioindicators: Approaches and Techniques for the Detection, Analysis, and Validation of Novel Biomarkers of Diseases" Pharmaceutics 15, no. 6: 1630. https://doi.org/10.3390/pharmaceutics15061630





