Porcine Mandibular Bone Marrow-Derived Mesenchymal Stem Cell (BMSC)-Derived Extracellular Vesicles Can Promote the Osteogenic Differentiation Capacity of Porcine Tibial-Derived BMSCs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.1.1. Gene Expression Omnibus (GEO) Database and Gene Microarray
2.1.2. Differentially Expressed Genes (DE Genes) and Target Genes
2.1.3. Enrichment, Gene Ontology, and Pathway of the Target Genes
2.1.4. Protein–Protein Interaction Network and Gene–Gene Network Construction
2.1.5. Gene Set Enrichment Analysis for the TLR4/NF-κB Signaling Pathway
2.2. Verification of the Results via Quantitative Polymerase Chain Reaction and Western Blot
Isolation and Cultivation of BMSCs
2.3. Isolation of Mandibular BMSC-Derived EVs
2.4. Characterization of Harvested mBMSC-EVs
2.4.1. Nanoparticle Tracking Analysis Using NanoSight NS300
2.4.2. Transmission Electron Microscopy
2.4.3. CD9, CD63, and CD81 Fluorescein Labeling of EVs
2.5. Cellular Uptake and Internalization of mBMSC-EVs
2.6. Verification of BMSCs’ Gene Expression Using qPCR
2.7. Verification of the Protein Expression Level Using Western Blot
2.8. Osteogenic Differentiation and Osteogenic Capacity Validation
2.8.1. ALP Staining
2.8.2. ARS Staining
2.8.3. Von Kossa Staining
2.9. Statistical Analysis
3. Results
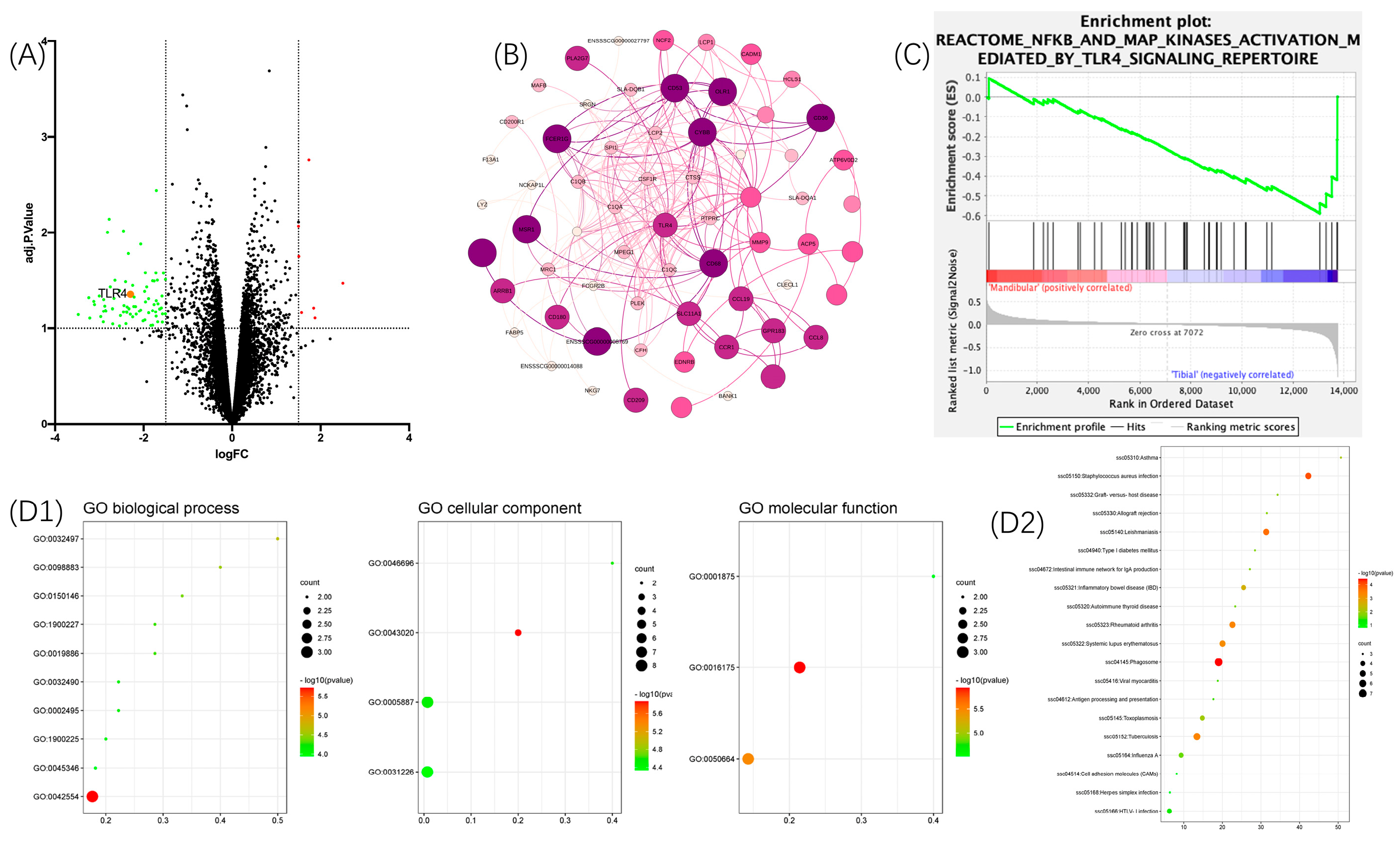
3.1. Bioinformatics Analysis—DE Genes
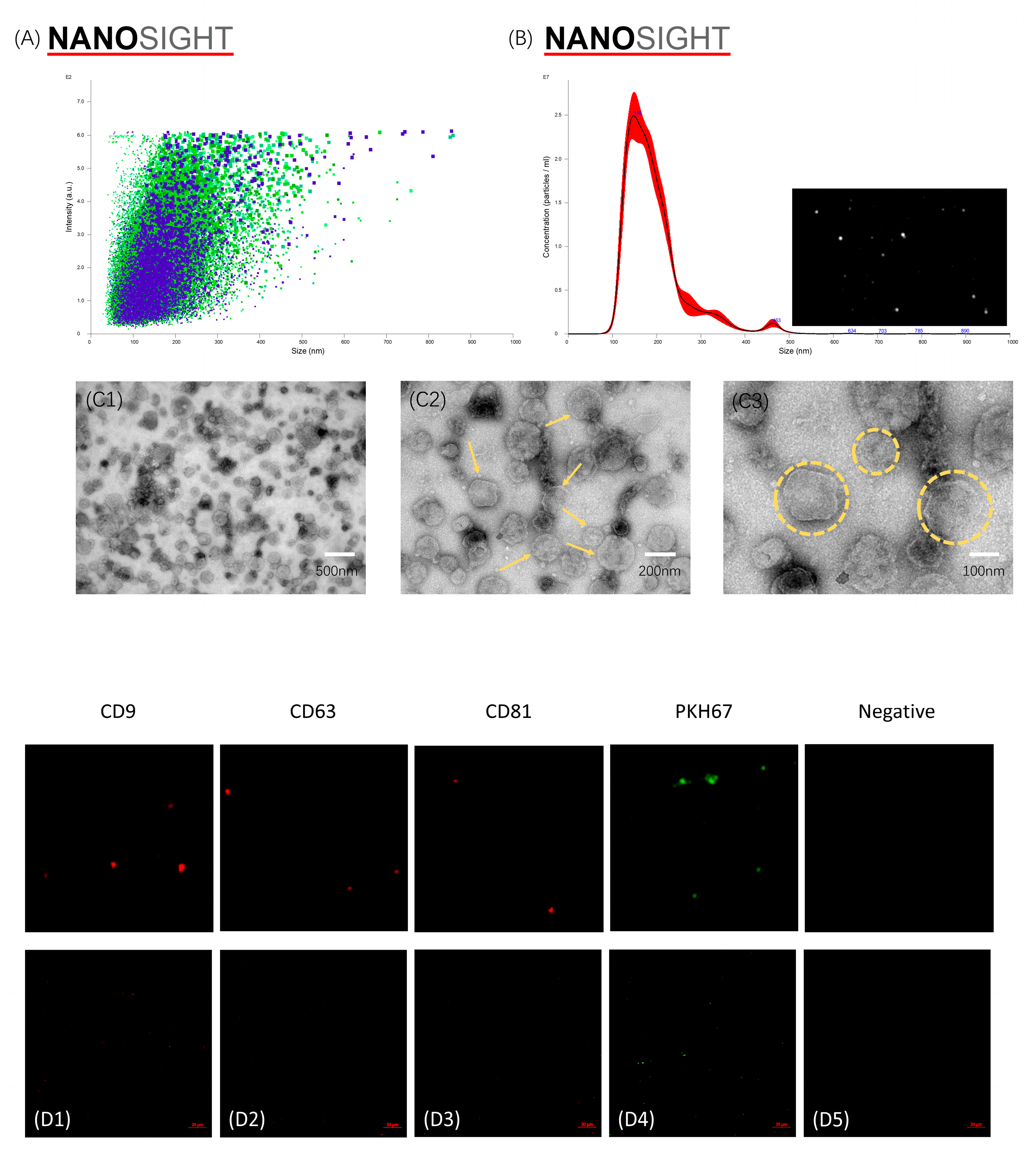
3.2. Characterization of mBMSC-EVs
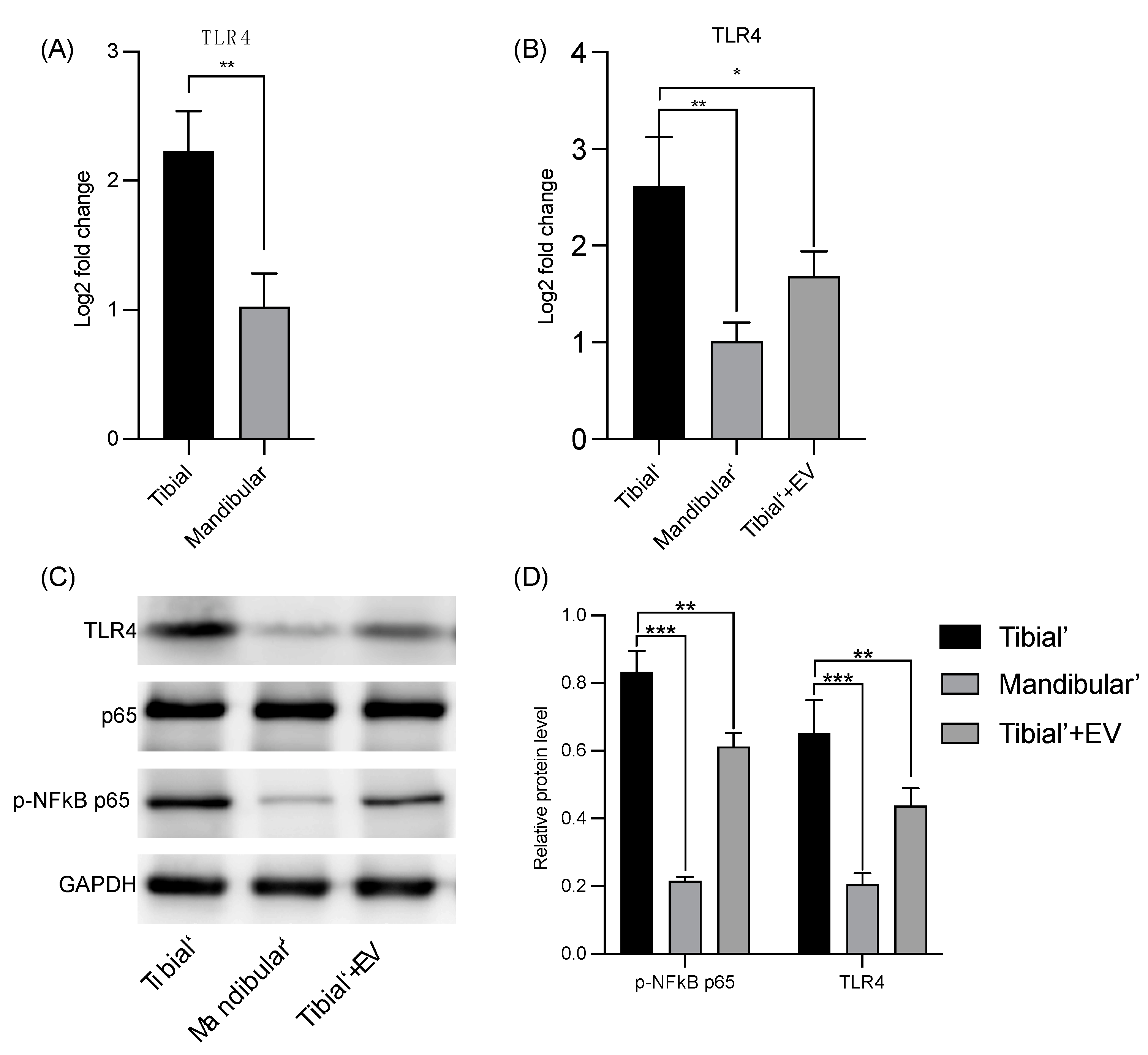
3.3. Verification of the Databank-Based Predicted Expression of TLR4 Using RT-qPCR and WB
3.4. Cellular Uptake of mBMSC-EVs
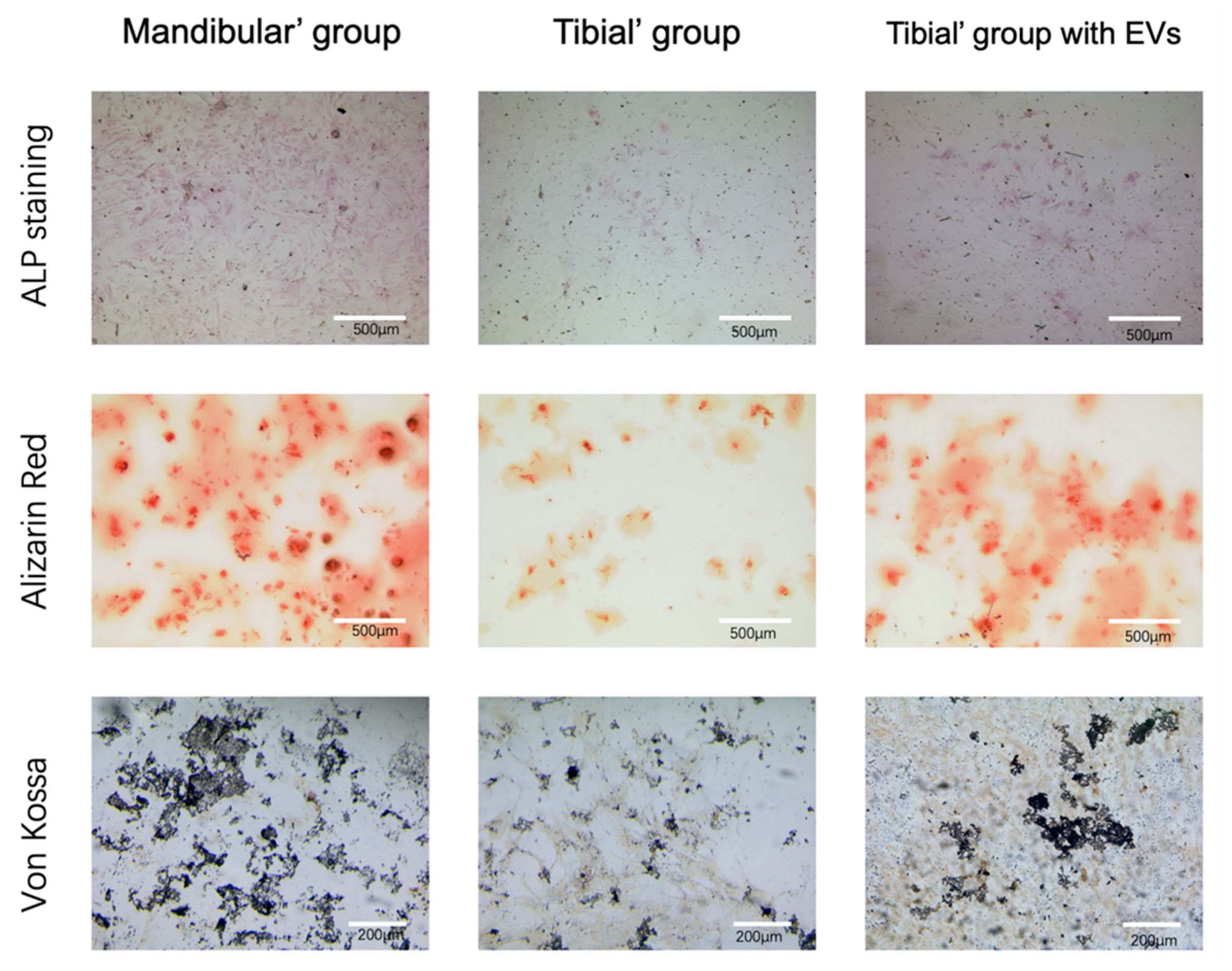
3.5. Verification of the Osteogenic Capacity of BMSCs (ALP/ARS/Von Kossa)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ekegren, C.; Edwards, E.; de Steiger, R.; Gabbe, B. Incidence, Costs and Predictors of Non-Union, Delayed Union and Mal-Union Following Long Bone Fracture. Int. J. Environ. Res. Public Health 2018, 15, 2845. [Google Scholar] [CrossRef]
- Hak, D.J.; Fitzpatrick, D.; Bishop, J.A.; Marsh, J.L.; Tilp, S.; Schnettler, R.; Simpson, H.; Alt, V. Delayed union and nonunions: Epidemiology, clinical issues, and financial aspects. Injury 2014, 45, S3–S7. [Google Scholar] [CrossRef]
- Bez, M.; Sheyn, D.; Tawackoli, W.; Avalos, P.; Shapiro, G.; Giaconi, J.C.; Da, X.; David, S.B.; Gavrity, J.; Awad, H.A.; et al. In situ bone tissue engineering via ultrasound-mediated gene delivery to endogenous progenitor cells in mini-pigs. Sci. Transl. Med. 2017, 9, eaal3128. [Google Scholar] [CrossRef]
- Hierholzer, C.; Sama, D.; Toro, J.B.; Peterson, M.; Helfet, D.L. Plate Fixation of Ununited Humeral Shaft Fractures: Effect of Type of Bone Graft on Healing. J. Bone Jt. Surg. 2006, 88, 1442–1447. [Google Scholar] [CrossRef]
- Younger, E.M.; Chapman, M.W. Morbidity at Bone Graft Donor Sites. J. Orthop. Trauma 1989, 3, 192–195. [Google Scholar] [CrossRef]
- Bosch-Rué, E.; Diez-Tercero, L.; Giordano-Kelhoffer, B.; Delgado, L.M.; Bosch, B.M.; Hoyos-Nogués, M.; Mateos-Timoneda, M.A.; Tran, P.A.; Gil, F.J.; Perez, R.A. Biological Roles and Delivery Strategies for Ions to Promote Osteogenic Induction. Front. Cell Dev. Biol. 2021, 8, 614545. [Google Scholar] [CrossRef]
- Behr, B.; Tang, C.; Germann, G.; Longaker, M.T.; Quarto, N. Locally Applied Vascular Endothelial Growth Factor A Increases the Osteogenic Healing Capacity of Human Adipose-Derived Stem Cells by Promoting Osteogenic and Endothelial Differentiation. Stem Cells 2011, 29, 286–296. [Google Scholar] [CrossRef]
- Lin, X.; Yang, S.; Lai, K.; Yang, H.; Webster, T.J.; Yang, L. Orthopedic implant biomaterials with both osteogenic and anti-infection capacities and associated in vivo evaluation methods. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J. Marrow Stromal Cells as Stem Cells for Nonhematopoietic Tissues. Science 1997, 276, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.J.; Chailakhyan, R.K.; Latsinik, N.V.; Panasyuk, A.F.; Keiliss-Borok, I.V. STROMAL CELLS RESPONSIBLE FOR TRANSFERRING THE MICROENVIRONMENT OF THE HEMOPOIETIC TISSUES: Cloning In Vitro and Retransplantation In Vivo. Transplantation 1974, 17, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Aghaloo, T.L.; Chaichanasakul, T.; Bezouglaia, O.; Kang, B.; Franco, R.; Dry, S.M.; Atti, E.; Tetradis, S. Osteogenic Potential of Mandibular vs. Long-bone Marrow Stromal Cells. J. Dent. Res. 2010, 89, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Ge, J.; Zhang, P.; Fu, Y.; Zhang, Z.; Cheng, J.; Jiang, H. Phenotypic characterization of craniofacial bone marrow stromal cells: Unique properties of enhanced osteogenesis, cell recruitment, autophagy, and apoptosis resistance. Cell Tissue Res. 2014, 358, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Akintoye, S.O.; Lam, T.; Shi, S.; Brahim, J.; Collins, M.T.; Robey, P.G. Skeletal site-specific characterization of orofacial and iliac crest human bone marrow stromal cells in same individuals. Bone 2006, 38, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE 2016, 11, e0148407. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- van Balkom, B.W.M.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Beer Stolz, D.; Sullivan, M.L.G.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Pishavar, E.; Copus, J.S.; Atala, A.; Lee, S.J. Comparison Study of Stem Cell-Derived Extracellular Vesicles for Enhanced Osteogenic Differentiation. Tissue Eng. Part A 2021, 27, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Lee, J.; Kim, H.K.; Yeom, S.H.; Woo, C.H.; Jung, Y.J.; Yun, Y.E.; Park, S.Y.; Han, J.; Kim, E.; et al. Extracellular vesicles from adipose tissue-derived stem cells alleviate osteoporosis through osteoprotegerin and miR-21-5p. J. Extracell. Vesicles 2021, 10, e12152. [Google Scholar] [CrossRef] [PubMed]
- Zhai, M.; Zhu, Y.; Yang, M.; Mao, C. Human Mesenchymal Stem Cell Derived Exosomes Enhance Cell-Free Bone Regeneration by Altering Their miRNAs Profiles. Adv. Sci. 2020, 7, 2001334. [Google Scholar] [CrossRef]
- Yahao, G.; Xinjia, W. The Role and Mechanism of Exosomes from Umbilical Cord Mesenchymal Stem Cells in Inducing Osteogenesis and Preventing Osteoporosis. Cell Transplant. 2021, 30, 096368972110574. [Google Scholar] [CrossRef]
- Lloyd, B.; Tee, B.C.; Headley, C.; Emam, H.; Mallery, S.; Sun, Z. Similarities and differences between porcine mandibular and limb bone marrow mesenchymal stem cells. Arch. Oral Biol. 2017, 77, 1–11. [Google Scholar] [CrossRef]
- AlQranei, M.S.; Senbanjo, L.T.; Aljohani, H.; Hamza, T.; Chellaiah, M.A. Lipopolysaccharide- TLR-4 Axis regulates Osteoclastogenesis independent of RANKL/RANK signaling. BMC Immunol. 2021, 22, 23. [Google Scholar] [CrossRef] [PubMed]
- Muthukuru, M.; Darveau, R.P. TLR signaling that induces weak inflammatory response and SHIP1 enhances osteogenic functions. Bone Res. 2014, 2, 14031. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, Y.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Bianco, P.; Kuznetsov, S.A.; Riminucci, M.; Gehron Robey, P. Postnatal Skeletal Stem Cells. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2006; Volume 419, pp. 117–148. ISBN 978-0-12-373650-5. [Google Scholar]
- Minakaki, G.; Menges, S.; Kittel, A.; Emmanouilidou, E.; Schaeffner, I.; Barkovits, K.; Bergmann, A.; Rockenstein, E.; Adame, A.; Marxreiter, F.; et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 2018, 14, 98–119. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Gregory, C.A.; Grady Gunn, W.; Peister, A.; Prockop, D.J. An Alizarin red-based assay of mineralization by adherent cells in culture: Comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004, 329, 77–84. [Google Scholar] [CrossRef]
- McNeill, E.P.; Zeitouni, S.; Pan, S.; Haskell, A.; Cesarek, M.; Tahan, D.; Clough, B.H.; Krause, U.; Dobson, L.K.; Garcia, M.; et al. Characterization of a pluripotent stem cell-derived matrix with powerful osteoregenerative capabilities. Nat. Commun. 2020, 11, 3025. [Google Scholar] [CrossRef]
- Jia, D.; Heersche, J.N.M. Insulin-like growth factor-1 and -2 stimulate osteoprogenitor proliferation and differentiation and adipocyte formation in cell populations derived from adult rat bone. Bone 2000, 27, 785–794. [Google Scholar] [CrossRef]
- Wang, D.; Gilbert, J.R.; Taylor, G.M.; Sodhi, C.P.; Hackam, D.J.; Losee, J.E.; Billiar, T.R.; Cooper, G.M. TLR4 Inactivation in Myeloid Cells Accelerates Bone Healing of a Calvarial Defect Model in Mice. Plast. Reconstr. Surg. 2017, 140, 296e–306e. [Google Scholar] [CrossRef]
- Zhu, D.; Yu, H.; Liu, P.; Yang, Q.; Chen, Y.; Luo, P.; Zhang, C.; Gao, Y. Calycosin modulates inflammation via suppressing TLR4/NF-κB pathway and promotes bone formation to ameliorate glucocorticoid-induced osteonecrosis of the femoral head in rat. Phytother. Res. 2021, 35, 2824–2835. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Bonsergent, E.; Grisard, E.; Buchrieser, J.; Schwartz, O.; Théry, C.; Lavieu, G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021, 12, 1864. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhang, Y.; Liu, H.; Cheng, Q.; Yang, S.; Yang, D. All-trans retinoic acid inhibits the osteogenesis of periodontal ligament stem cells by promoting IL-1β production via NF-κB signaling. Int. Immunopharmacol. 2022, 108, 108757. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Fan, L.; Nan, K.; Li, J.; Shi, Z.; Dang, X.; Wang, K. Excessive Activation of TLR4/NF-κB Interactively Suppresses the Canonical Wnt/β-catenin Pathway and Induces SANFH in SD Rats. Sci. Rep. 2017, 7, 11928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cao, H.; Guo, J.; Yuan, Y.; Ni, G. Effects of BMSC-Derived EVs on Bone Metabolism. Pharmaceutics 2022, 14, 1012. [Google Scholar] [CrossRef]
- Hu, H.; Wang, D.; Li, L.; Yin, H.; He, G.; Zhang, Y. Role of microRNA-335 carried by bone marrow mesenchymal stem cells-derived extracellular vesicles in bone fracture recovery. Cell Death Dis. 2021, 12, 156. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, S.D.; Kook, M.; Lee, H.Y.; Ghim, J.; Choi, Y.; Zabel, B.A.; Ryu, S.H.; Bae, Y.-S. Phospholipase D2 drives mortality in sepsis by inhibiting neutrophil extracellular trap formation and down-regulating CXCR2. J. Exp. Med. 2015, 212, 1381–1390. [Google Scholar] [CrossRef]
- Christian, F.; Smith, E.; Carmody, R. The Regulation of NF-κB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Li, R.; Qing, Y.; Ying, B.; Qin, Y. Biomaterial-based osteoimmunomodulatory strategies via the TLR4-NF-κB signaling pathway: A review. Appl. Mater. Today 2021, 22, 100969. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, B.; Zhu, W.; Shi, J.; Wang, Y.; Si, M. IL-1 Receptor Antagonist Protects the Osteogenesis Capability of Gingival-Derived Stem/Progenitor Cells under Inflammatory Microenvironment Induced by Porphyromonas gingivalis Lipopolysaccharides. Stem Cells Int. 2021, 2021, 6638575. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Qiao, S.; Wang, W.; Zhang, Y.; Shi, J.; Zhang, X.; Gu, W.; Zhang, X.; Li, Y.; Ding, X.; et al. Melatonin prevents peri-implantitis via suppression of TLR4/NF-κB. Acta Biomater. 2021, 134, 325–336. [Google Scholar] [CrossRef]
- Zheng, L.; Shen, X.; Ye, J.; Xie, Y.; Yan, S. Metformin alleviates hyperglycemia-induced apoptosis and differentiation suppression in osteoblasts through inhibiting the TLR4 signaling pathway. Life Sci. 2019, 216, 29–38. [Google Scholar] [CrossRef]
- Wang, C.; Wang, W.; Jin, X.; Shen, J.; Hu, W.; Jiang, T. Puerarin attenuates inflammation and oxidation in mice with collagen antibody-induced arthritis via TLR4/NF-κB signaling. Mol. Med. Rep. 2016, 14, 1365–1370. [Google Scholar] [CrossRef]
- Potter, M.L.; Smith, K.; Vyavahare, S.; Kumar, S.; Periyasamy-Thandavan, S.; Hamrick, M.; Isales, C.M.; Hill, W.D.; Fulzele, S. Characterization of Differentially Expressed miRNAs by CXCL12/SDF-1 in Human Bone Marrow Stromal Cells. Biomol. Concepts 2021, 12, 132–143. [Google Scholar] [CrossRef]
- Essandoh, K.; Fan, G.-C. Role of extracellular and intracellular microRNAs in sepsis. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2014, 1842, 2155–2162. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Zhao, H.; Han, X.; Zhao, T.; Qu, P.; Li, G.; Wang, W. Extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cell promotes osteogenic differentiation via FTO inhibition. Stem Cell Res. Ther. 2020, 11, 227. [Google Scholar] [CrossRef]
- Wang, X.; Zou, C.; Hou, C.; Bian, Z.; Jiang, W.; Li, M.; Zhu, L. Extracellular vesicles from bone marrow mesenchymal stem cells alleviate osteoporosis in mice through USP7-mediated YAP1 protein stability and the Wnt/β-catenin pathway. Biochem. Pharmacol. 2023, 217, 115829. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Xie, H.; Jin, Q.; Zhao, X.; Shi, Y.; Zhou, Y.; Hu, Z.; Ye, Y.; Huang, X.; Sun, Y.; et al. Extracellular vesicles derived from neural EGFL-Like 1-modified mesenchymal stem cells improve acellular bone regeneration via the miR-25-5p-SMAD2 signaling axis. Bioact. Mater. 2022, 17, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Changani, R.; Gagal, K.; Bhargava, R.; Sankhla, S. Percutaneous autologus bone marrow injection in the treatment of delayed or nonunion. Indian J. Orthop. 2007, 41, 67. [Google Scholar] [CrossRef] [PubMed]
- Emara, K.M. Recent biological trends in management of fracture non-union. World J. Orthop. 2015, 6, 623. [Google Scholar] [CrossRef]

| Gene | Accession Number | Primer Sequences |
|---|---|---|
| TLR4 | NM_001113039.2 | Forward: 5′-CTGCCTTCACTACAGAGACTTCATTCC-3′ |
| Reverse: 5′-CACCACGACAATAACCTTCCGACTT-3′ | ||
| GAPDH | NM_001206359.1 | Forward: 5′-GTGAAGGTCGGAGTGAACGGATT-3′ |
| Reverse: 5′-ACCATGTAGTGGAGGTCAATGAAGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Zhang, X.; Li, Y.; He, Z.; Qin, K.; Buhl, E.M.; Mert, Ü.; Horst, K.; Hildebrand, F.; Balmayor, E.R.; et al. Porcine Mandibular Bone Marrow-Derived Mesenchymal Stem Cell (BMSC)-Derived Extracellular Vesicles Can Promote the Osteogenic Differentiation Capacity of Porcine Tibial-Derived BMSCs. Pharmaceutics 2024, 16, 279. https://doi.org/10.3390/pharmaceutics16020279
Zhao Q, Zhang X, Li Y, He Z, Qin K, Buhl EM, Mert Ü, Horst K, Hildebrand F, Balmayor ER, et al. Porcine Mandibular Bone Marrow-Derived Mesenchymal Stem Cell (BMSC)-Derived Extracellular Vesicles Can Promote the Osteogenic Differentiation Capacity of Porcine Tibial-Derived BMSCs. Pharmaceutics. 2024; 16(2):279. https://doi.org/10.3390/pharmaceutics16020279
Chicago/Turabian StyleZhao, Qun, Xing Zhang, You Li, Zhizhen He, Kang Qin, Eva Miriam Buhl, Ümit Mert, Klemens Horst, Frank Hildebrand, Elizabeth R. Balmayor, and et al. 2024. "Porcine Mandibular Bone Marrow-Derived Mesenchymal Stem Cell (BMSC)-Derived Extracellular Vesicles Can Promote the Osteogenic Differentiation Capacity of Porcine Tibial-Derived BMSCs" Pharmaceutics 16, no. 2: 279. https://doi.org/10.3390/pharmaceutics16020279





