Immune Specific and Tumor-Dependent mRNA Vaccines for Cancer Immunotherapy: Reprogramming Clinical Translation into Tumor Editing Therapy
Abstract
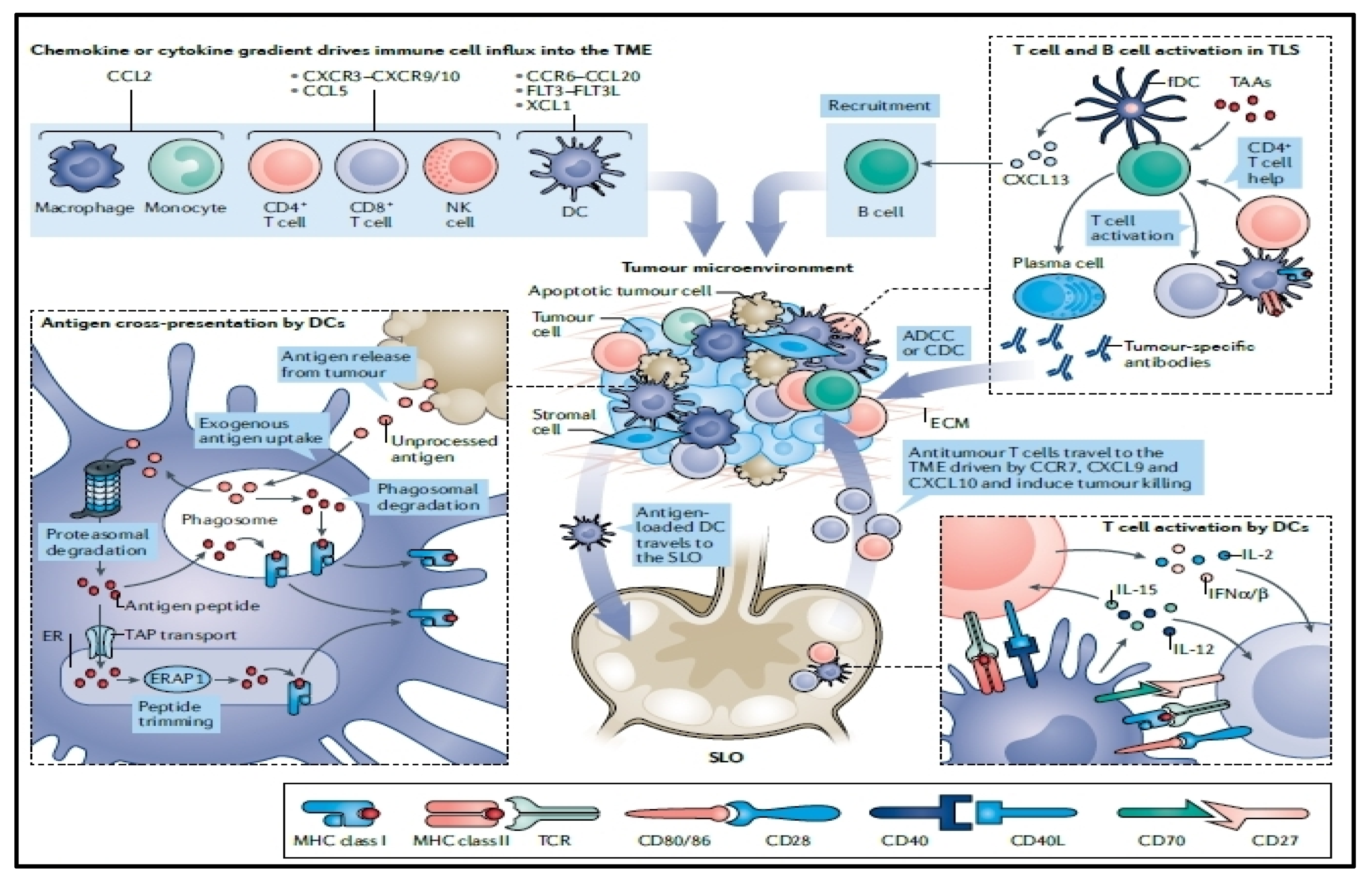
:1. Introduction
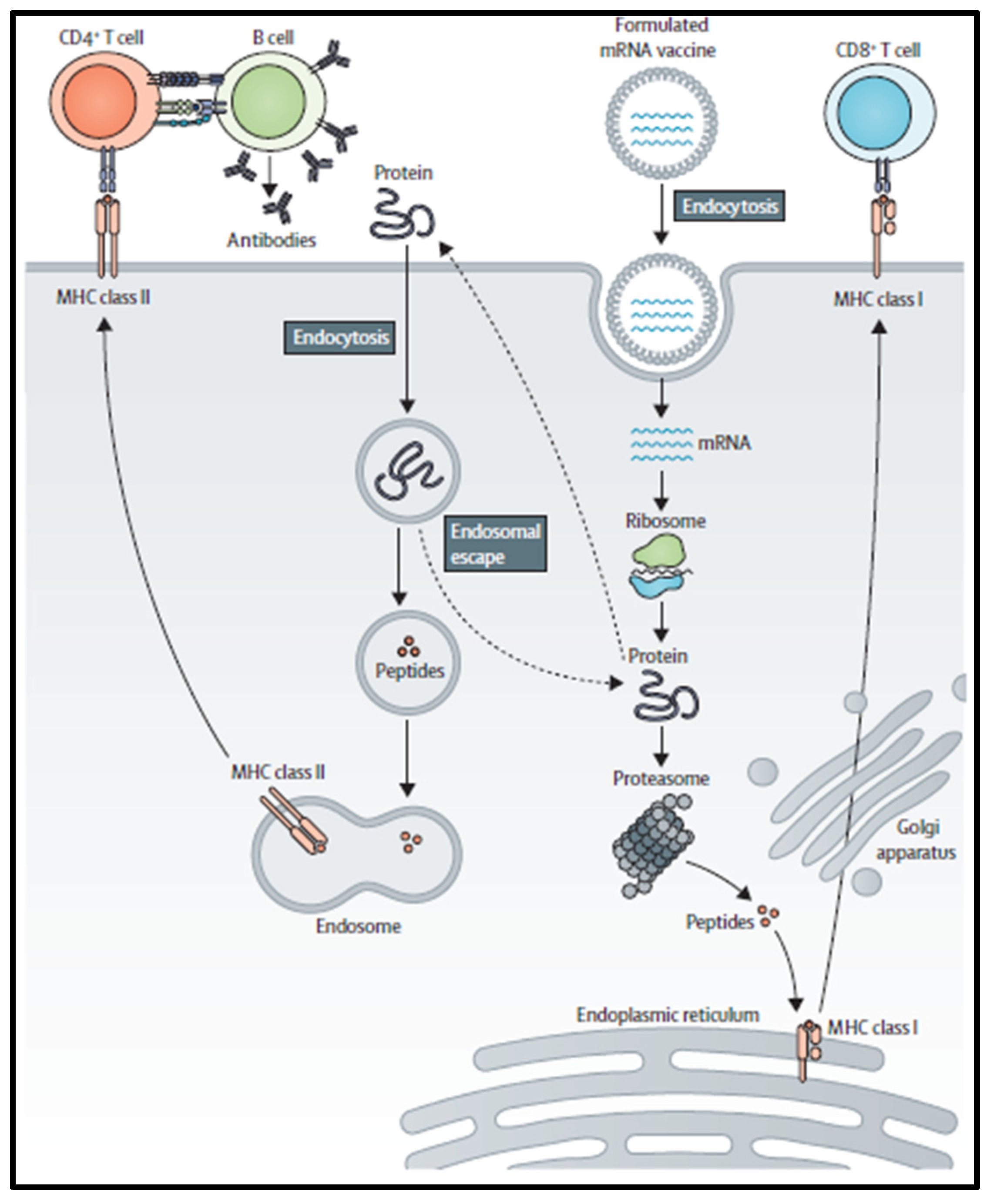
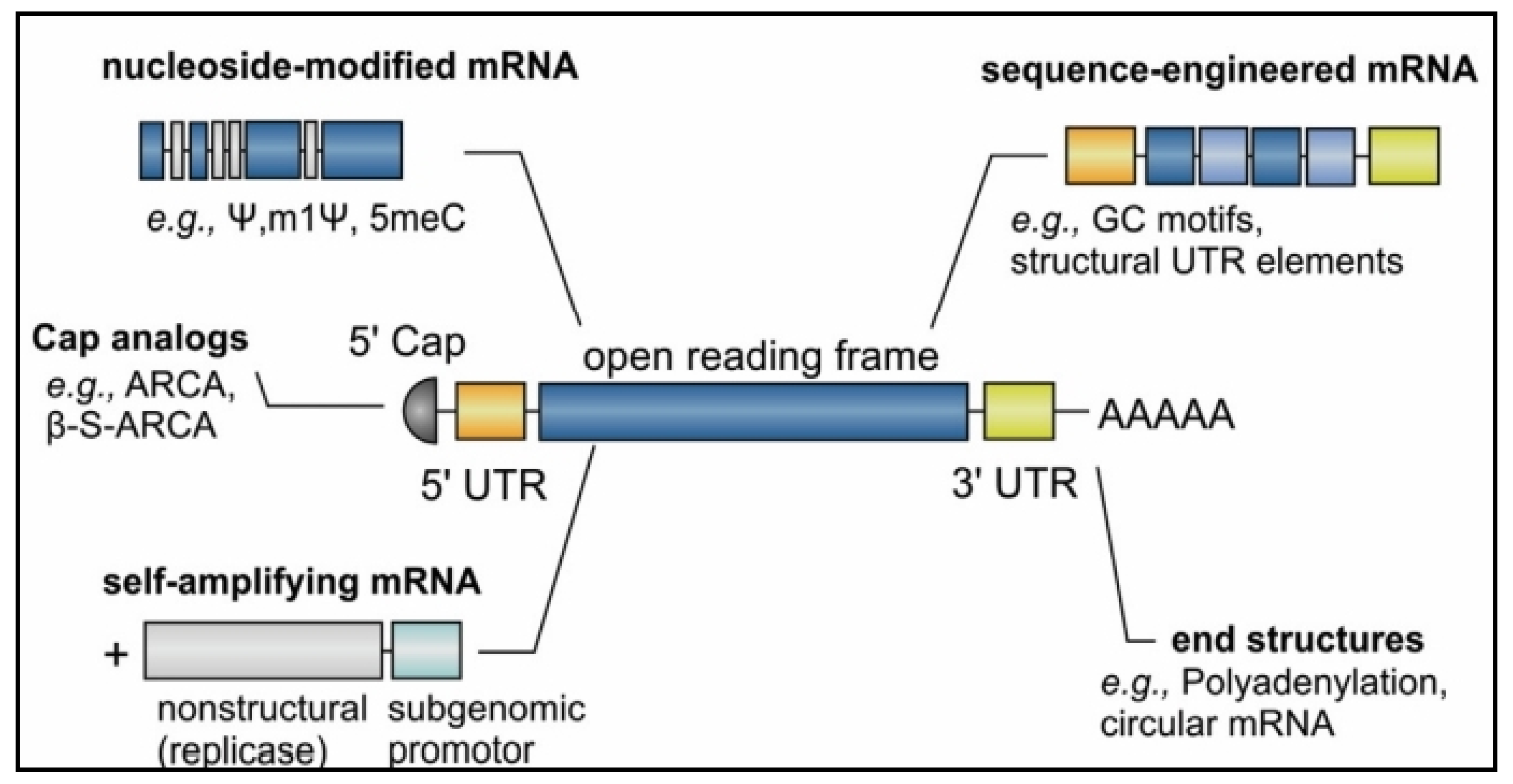
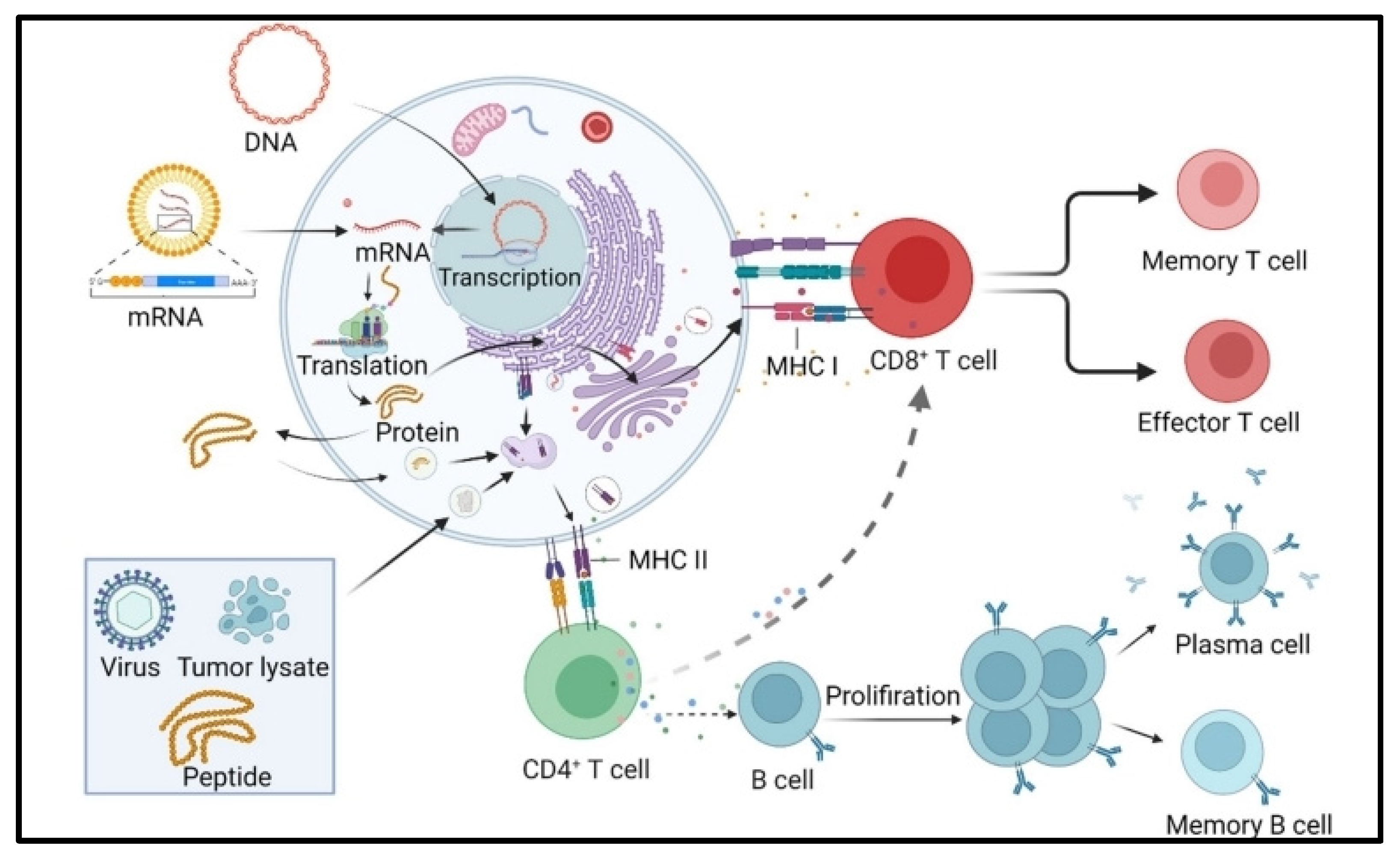
2. mRNA Vaccine Pharmacology
3. Optimization of mRNA Translation
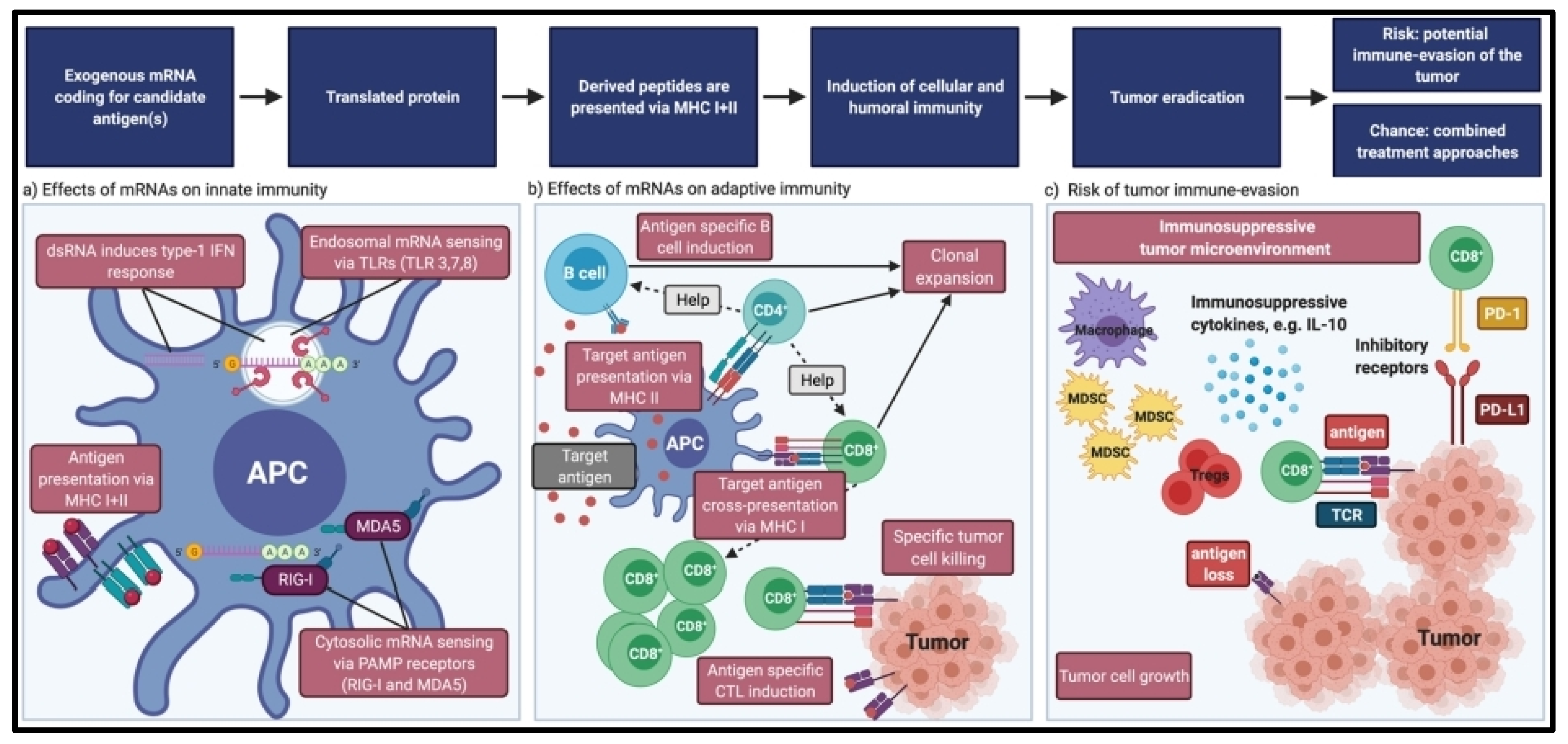
4. Immunogenicity of mRNA Vaccines
5. mRNA Vaccine Efficacy
6. Tumor Intrinsic Resistance
7. Tumor Extrinsic Resistance
8. Cell-Based Cancer Vaccines
9. Peptide-Based Cancer Vaccines
10. Nucleic Acid-Based Cancer Vaccines
11. Viral-Based Cancer Vaccines
12. Lipid mRNA-Based Nanovaccines
13. Dendritic Cell mRNA-Based Vaccines
14. Challenges and Future Perspectives
15. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Szabó, G.T.; Mahiny, A.J.; Vlatkovic, I. COVID-19 mRNA vaccines: Platforms and current developments. Mol. Ther. 2022, 30, 1850–1868. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Lee, S.S. From COVID-19 to Cancer mRNA Vaccines: Moving from Bench to Clinic in the Vaccine Landscape. Front. Immunol. 2021, 12, 679344. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Verbeke, R.; Hogan, M.J.; Loré, K.; Pardi, N. Innate immune mechanisms of mRNA vaccines. Immunity 2022, 55, 1993–2005. [Google Scholar] [CrossRef]
- Lorentzen, C.L.; Haanen, J.B.; Met, Ö.; Svane, I.M. Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol. 2022, 23, e450–e458. [Google Scholar] [CrossRef]
- Adamik, J.; Butterfield, L.H. What’s next for cancer vaccines. Sci. Transl. Med. 2022, 14, eabo4632. [Google Scholar] [CrossRef]
- Lopes, A.; Vandermeulen, G.; Préat, V. Cancer DNA vaccines: Current preclinical and clinical developments and future perspectives. J. Exp. Clin. Cancer Res. 2019, 38, 146. [Google Scholar] [CrossRef]
- Liang, W.; Lin, Z.; Du, C.; Qiu, D.; Zhang, Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol. Cancer 2020, 19, 38. [Google Scholar] [CrossRef]
- Jia, L.; Mao, Y.; Ji, Q.; Dersh, D.; Yewdell, J.W.; Qian, S.B. Decoding mRNA translatability and stability from the 5’ UTR. Nat. Struct. Mol. Biol. 2020, 27, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Presnyak, V.; Alhusaini, N.; Chen, Y.H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Bidram, M.; Zhao, Y.; Shebardina, N.G.; Baldin, A.V.; Bazhin, A.V.; Ganjalikhany, M.R.; Zamyatnin, A.A., Jr. Ganjalikhani-Hakemi, M. mRNA-Based Cancer Vaccines: A Therapeutic Strategy for the Treatment of Melanoma Patients. Vaccines 2021, 9, 1060. [Google Scholar] [CrossRef]
- Liu, C.; Shi, Q.; Huang, X.; Koo, S.; Kong, N.; Tao, W. mRNA-based cancer therapeutics. Nat. Rev. Cancer 2023, 23, 526–543. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. Three decades of messenger RNA vaccine development. Nano Today 2019, 28, 100766. [Google Scholar] [CrossRef]
- Huang, X.; Kong, N.; Zhang, X.; Cao, Y.; Langer, R.; Tao, W. The landscape of mRNA nanomedicine. Nat. Med. 2022, 28, 2273–2287. [Google Scholar] [CrossRef]
- Heine, A.; Juranek, S.; Brossart, P. Clinical and immunological effects of mRNA vaccines in malignant diseases. Mol. Cancer 2021, 20, 52. [Google Scholar] [CrossRef]
- Fu, J.; Chen, F.; Lin, Y.; Gao, J.; Chen, A.; Yang, J. Discovery and characterization of tumor antigens in hepatocellular carcinoma for mRNA vaccine development. J. Cancer Res. Clin. Oncol. 2023, 149, 4047–4061. [Google Scholar] [CrossRef]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Rohner, E.; Yang, R.; Foo, K.S.; Goedel, A.; Chien, K.R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 2022, 40, 1586–1600. [Google Scholar] [CrossRef]
- Bae, H.; Coller, J. Codon optimality-mediated mRNA degradation: Linking translational elongation to mRNA stability. Mol. Cell 2022, 82, 1467–1476. [Google Scholar] [CrossRef]
- Schlake, T.; Thran, M.; Fiedler, K.; Heidenreich, R.; Petsch, B.; Fotin-Mleczek, M. mRNA: A Novel Avenue to Antibody Therapy? Mol. Ther. 2019, 27, 773–784. [Google Scholar] [CrossRef]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell. Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Eralp, Y. Application of mRNA Technology in Cancer Therapeutics. Vaccines 2022, 10, 1262. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, E.; Boczkowski, D.; Nair, S.K. The Quest for mRNA Vaccines. Nucleic Acid Ther. 2022, 32, 449–456. [Google Scholar] [CrossRef]
- Ye, Z.; Harmon, J.; Ni, W.; Li, Y.; Wich, D.; Xu, Q. The mRNA Vaccine Revolution: COVID-19 Has Launched the Future of Vaccinology. ACS Nano 2023, 17, 15231–15253. [Google Scholar] [CrossRef]
- Blanchard, E.L.; Loomis, K.H.; Bhosle, S.M.; Vanover, D.; Baumhof, P.; Pitard, B.; Zurla, C.; Santangelo, P.J. Proximity Ligation Assays for In Situ Detection of Innate Immune Activation: Focus on In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 14, 52–66. [Google Scholar] [CrossRef]
- Rosini, R.; Nicchi, S.; Pizza, M.; Rappuoli, R. Vaccines against Antimicrobial Resistance. Front. Immunol. 2020, 11, 1048. [Google Scholar] [CrossRef]
- Klugman, K.P.; Black, S. Impact of existing vaccines in reducing antibiotic resistance: Primary and secondary effects. Proc. Natl. Acad. Sci. USA 2018, 115, 12896–12901. [Google Scholar] [CrossRef]
- van Elsas, M.J.; van Hall, T.; van der Burg, S.H. Future Challenges in Cancer Resistance to Immunotherapy. Cancers 2020, 12, 935. [Google Scholar] [CrossRef]
- Zhou, J.; Ji, Q.; Li, Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: Underlying mechanisms and reversal strategies. J. Exp. Clin. Cancer Res. 2021, 40, 328. [Google Scholar] [CrossRef] [PubMed]
- Kirchhammer, N.; Trefny, M.P.; Auf der Maur, P.; Läubli, H.; Zippelius, A. Combination cancer immunotherapies: Emerging treatment strategies adapted to the tumor microenvironment. Sci. Transl. Med. 2022, 14, eabo3605. [Google Scholar] [CrossRef]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021, 184, 5309–5337. [Google Scholar] [CrossRef]
- Pitt, J.M.; Vétizou, M.; Daillère, R.; Roberti, M.P.; Yamazaki, T.; Routy, B.; Lepage, P.; Boneca, I.G.; Chamaillard, M.; Kroemer, G.; et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016, 44, 1255–1269. [Google Scholar] [CrossRef]
- Valeri, A.; García-Ortiz, A.; Castellano, E.; Córdoba, L.; Maroto-Martín, E.; Encinas, J.; Leivas, A.; Río, P.; Martínez-López, J. Overcoming tumor resistance mechanisms in CAR-NK cell therapy. Front. Immunol. 2022, 13, 953849. [Google Scholar] [CrossRef]
- Barry, S.T.; Gabrilovich, D.I.; Sansom, O.J.; Campbell, A.D.; Morton, J.P. Therapeutic targeting of tumour myeloid cells. Nat. Rev. Cancer 2023, 23, 216–237. [Google Scholar] [CrossRef]
- Saw, P.E.; Chen, J.; Song, E. Targeting CAFs to overcome anticancer therapeutic resistance. Trends Cancer 2022, 8, 527–555. [Google Scholar] [CrossRef]
- Kloosterman, D.J.; Akkari, L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell 2023, 186, 1627–1651. [Google Scholar] [CrossRef]
- Christofides, A.; Strauss, L.; Yeo, A.; Cao, C.; Charest, A.; Boussiotis, V.A. The complex role of tumor-infiltrating macrophages. Nat. Immunol. 2022, 23, 1148–1156. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Sellars, M.C.; Wu, C.J.; Fritsch, E.F. Cancer vaccines: Building a bridge over troubled waters. Cell 2022, 185, 2770–2788. [Google Scholar] [CrossRef]
- Harari, A.; Graciotti, M.; Bassani-Sternberg, M.; Kandalaft, L.E. Antitumour dendritic cell vaccination in a priming and boosting approach. Nat. Rev. Drug Discov. 2020, 19, 635–652. [Google Scholar] [CrossRef]
- Palucka, K.; Banchereau, J. Dendritic-cell-based therapeutic cancer vaccines. Immunity 2013, 39, 38–48. [Google Scholar] [CrossRef]
- Sabado, R.L.; Balan, S.; Bhardwaj, N. Dendritic cell-based immunotherapy. Cell Res. 2017, 27, 74–95. [Google Scholar] [CrossRef]
- Gardner, A.; de Mingo Pulido, Á.; Ruffell, B. Dendritic Cells and Their Role in Immunotherapy. Front. Immunol. 2020, 11, 924. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Zhou, L.; Mi, Q.S.; Jiang, A. Plasmacytoid Dendritic Cells and Cancer Immunotherapy. Cells 2022, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.J.; Burgess-Brown, N.A.; Jiang, S. Beyond Just Peptide Antigens: The Complex World of Peptide-Based Cancer Vaccines. Front. Immunol. 2021, 12, 696791. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef]
- Kimura, T.; Egawa, S.; Uemura, H. Personalized peptide vaccines and their relation to other therapies in urological cancer. Nat. Rev. Urol. 2017, 14, 501–510. [Google Scholar] [CrossRef]
- Parmiani, G.; Castelli, C.; Dalerba, P.; Mortarini, R.; Rivoltini, L.; Marincola, F.M.; Anichini, A. Cancer immunotherapy with peptide-based vaccines: What have we achieved? Where are we going? J. Natl. Cancer Inst. 2002, 94, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; de Izarra, A.; Degrouard, J.; Olive, E.; Maiti, P.K.; Jang, Y.H.; Lansac, Y. Protamine-Controlled Reversible DNA Packaging: A Molecular Glue. ACS Nano 2021, 15, 13094–13104. [Google Scholar] [CrossRef]
- Ukogu, O.A.; Smith, A.D.; Devenica, L.M.; Bediako, H.; McMillan, R.B.; Ma, Y.; Balaji, A.; Schwab, R.D.; Anwar, S.; Dasgupta, M.; et al. Protamine loops DNA in multiple steps. Nucleic Acids Res. 2020, 48, 6108–6119. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.H.; Ghadimi, K.; Kizhakkedathu, J.N.; Iba, T. What’s fishy about protamine? Clinical use, adverse reactions, and potential alternatives. J. Thromb. Haemost. 2023, 21, 1714–1723. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.; Yakkundi, A.; McKeen, H.D.; McClements, L.; McKeogh, T.J.; McCrudden, C.M.; Arthur, K.; Robson, T.; McCarthy, H.O. RALA-mediated delivery of FKBPL nucleic acid therapeutics. Nanomedicine 2015, 10, 2989–3001. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, B.; Ruiz, E.F.; Zhang, F. Advances in the polymeric delivery of nucleic acid vaccines. Theranostics 2022, 12, 4081–4109. [Google Scholar] [CrossRef] [PubMed]
- Teplensky, M.H.; Evangelopoulos, M.; Dittmar, J.W.; Forsyth, C.M.; Sinegra, A.J.; Wang, S.; Mirkin, C.A. Multi-antigen spherical nucleic acid cancer vaccines. Nat. Biomed. Eng. 2023, 7, 911–927. [Google Scholar] [CrossRef]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef]
- Desmet, C.J.; Ishii, K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012, 12, 479–491. [Google Scholar] [CrossRef]
- Iurescia, S.; Fioretti, D.; Rinaldi, M. Targeting Cytosolic Nucleic Acid-Sensing Pathways for Cancer Immunotherapies. Front. Immunol. 2018, 9, 711. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, C.A.; Bartas, M.; Volná, A.; Pečinka, P.; Blundell, T.L. Are There Hidden Genes in DNA/RNA Vaccines? Front. Immunol. 2022, 13, 801915. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Luo, J.; Han, X.; Wei, Y.; Wei, X. mRNA vaccine: A potential therapeutic strategy. Mol. Cancer 2021, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. The tangled history of mRNA vaccines. Nature 2021, 597, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, B.; Claerhout, S.; Carrasco, J.; Bar, I.; Corthals, J.; Wilgenhof, S.; Neyns, B.; Thielemans, K. TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: Link between T-cell activation and clinical responses in advanced melanoma. J. Immunother. Cancer 2020, 8, e000329. [Google Scholar] [CrossRef]
- Wang, S.; Liang, B.; Wang, W.; Li, L.; Feng, N.; Zhao, Y.; Wang, T.; Yan, F.; Yang, S.; Xia, X. Viral vectored vaccines: Design, development, preventive and therapeutic applications in human diseases. Signal Transduct. Target. Ther. 2023, 8, 149. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Drysdale, S.B.; Barr, R.S.; Rollier, C.S.; Green, C.A.; Pollard, A.J.; Sande, C.J. Priorities for developing respiratory syncytial virus vaccines in different target populations. Sci. Transl. Med. 2020, 12, eaax2466. [Google Scholar] [CrossRef]
- Sun, S.; Liu, Y.; He, C.; Hu, W.; Liu, W.; Huang, X.; Wu, J.; Xie, F.; Chen, C.; Wang, J.; et al. Combining NanoKnife with M1 oncolytic virus enhances anticancer activity in pancreatic cancer. Cancer Lett. 2021, 502, 9–24. [Google Scholar] [CrossRef]
- Soliman, H.; Hogue, D.; Han, H.; Mooney, B.; Costa, R.; Lee, M.C.; Niell, B.; Williams, A.; Chau, A.; Falcon, S.; et al. Oncolytic T-VEC virotherapy plus neoadjuvant chemotherapy in nonmetastatic triple-negative breast cancer: A phase 2 trial. Nat. Med. 2023, 29, 450–457. [Google Scholar] [CrossRef]
- Madan, R.A.; Bilusic, M.; Heery, C.; Schlom, J.; Gulley, J.L. Clinical evaluation of TRICOM vector therapeutic cancer vaccines. Semin. Oncol. 2012, 39, 296–304. [Google Scholar] [CrossRef]
- DeMaria, P.J.; Lee-Wisdom, K.; Donahue, R.N.; Madan, R.A.; Karzai, F.; Schwab, A.; Palena, C.; Jochems, C.; Floudas, C.; Strauss, J.; et al. Phase 1 open-label trial of intravenous administration of MVA-BN-brachyury-TRICOM vaccine in patients with advanced cancer. J. Immunother. Cancer 2021, 9, e003238. [Google Scholar] [CrossRef]
- Gregoriadis, G. Liposomes in Drug Delivery: How It All Happened. Pharmaceutics 2016, 8, 19. [Google Scholar] [CrossRef]
- Huang, T.; Peng, L.; Han, Y.; Wang, D.; He, X.; Wang, J.; Ou, C. Lipid nanoparticle-based mRNA vaccines in cancers: Current advances and future prospects. Front. Immunol. 2022, 13, 922301. [Google Scholar] [CrossRef]
- Chen, J.; Ye, Z.; Huang, C.; Qiu, M.; Song, D.; Li, Y.; Xu, Q. Lipid nanoparticle-mediated lymph node-targeting delivery of mRNA cancer vaccine elicits robust CD8+ T cell response. Proc. Natl. Acad. Sci. USA 2022, 119, e2207841119. [Google Scholar] [CrossRef]
- Poh, A. mRNA Vaccine Slows Melanoma Recurrence. Cancer Discov. 2023, 13, 1278. [Google Scholar] [CrossRef]
- Nagasaka, M.; Potugari, B.; Nguyen, A.; Sukari, A.; Azmi, A.S.; Ou, S.I. KRAS Inhibitors- yes but what next? Direct targeting of KRAS- vaccines, adoptive T cell therapy and beyond. Cancer Treat. Rev. 2021, 101, 102309. [Google Scholar] [CrossRef]
- Gilboa, E. DC-based cancer vaccines. J. Clin. Investig. 2007, 117, 1195–1203. [Google Scholar] [CrossRef]
- Cintolo, J.A.; Datta, J.; Mathew, S.J.; Czerniecki, B.J. Dendritic cell-based vaccines: Barriers and opportunities. Future Oncol. 2012, 8, 1273–1299. [Google Scholar] [CrossRef]
- Saxena, M.; van der Burg, S.H.; Melief, C.J.M.; Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer 2021, 21, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.A.; de Haas, A.M.; Geijtenbeek, T.B.H.; van Ree, R.; Tas, S.W.; van Kooyk, Y.; de Jong, E.C. Therapeutic Liposomal Vaccines for Dendritic Cell Activation or Tolerance. Front. Immunol. 2021, 12, 674048. [Google Scholar] [CrossRef]
- Mastelic-Gavillet, B.; Balint, K.; Boudousquie, C.; Gannon, P.O.; Kandalaft, L.E. Personalized Dendritic Cell Vaccines-Recent Breakthroughs and Encouraging Clinical Results. Front. Immunol. 2019, 10, 766. [Google Scholar] [CrossRef]
- Figlin, R.A.; Tannir, N.M.; Uzzo, R.G.; Tykodi, S.S.; Chen, D.Y.T.; Master, V.; Kapoor, A.; Vaena, D.; Lowrance, W.; Bratslavsky, G.; et al. Results of the ADAPT Phase 3 Study of Rocapuldencel-T in Combination with Sunitinib as First-Line Therapy in Patients with Metastatic Renal Cell Carcinoma. Clin. Cancer Res. 2020, 26, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- Vogelzang, N.J.; Beer, T.M.; Gerritsen, W.; Oudard, S.; Wiechno, P.; Kukielka-Budny, B.; Samal, V.; Hajek, J.; Feyerabend, S.; Khoo, V.; et al. Efficacy and Safety of Autologous Dendritic Cell-Based Immunotherapy, Docetaxel, and Prednisone vs. Placebo in Patients with Metastatic Castration-Resistant Prostate Cancer: The VIABLE Phase 3 Randomized Clinical Trial. JAMA Oncol. 2022, 8, 546–552. [Google Scholar] [CrossRef]
- Lee, J.B.; Chen, B.; Vasic, D.; Law, A.D.; Zhang, L. Cellular immunotherapy for acute myeloid leukemia: How specific should it be? Blood Rev. 2019, 35, 18–31. [Google Scholar] [CrossRef]
- Batich, K.A.; Mitchell, D.A.; Healy, P.; Herndon, J.E., 2nd; Sampson, J.H. Once, Twice, Three Times a Finding: Reproducibility of Dendritic Cell Vaccine Trials Targeting Cytomegalovirus in Glioblastoma. Clin. Cancer Res. 2020, 26, 5297–5303. [Google Scholar] [CrossRef]
- Sobhani, N.; Scaggiante, B.; Morris, R.; Chai, D.; Catalano, M.; Tardiel-Cyril, D.R.; Neeli, P.; Roviello, G.; Mondani, G.; Li, Y. Therapeutic cancer vaccines: From biological mechanisms and engineering to ongoing clinical trials. Cancer Treat. Rev. 2022, 109, 102429. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Chong, C.; Coukos, G.; Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 2022, 40, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Qu, H.; Wang, X.; Sobhani, N.; Wang, L.; Liu, S.; Xiong, W.; Zeng, Z.; Li, Y. Cancer/testis antigens: From serology to mRNA cancer vaccine. Semin. Cancer Biol. 2021, 76, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Benn, C.S.; Fisker, A.B.; Rieckmann, A.; Sørup, S.; Aaby, P. Vaccinology: Time to change the paradigm? Lancet Infect. Dis. 2020, 20, e274–e283. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Y.; Guo, M.; Du, S.; Han, N. Recent strategies for nano-based PTT combined with immunotherapy: From a biomaterial point of view. Theranostics 2021, 11, 7546–7569. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, C.S.; Hsu, J.C.; Hosseini, I.; Shen, B.Q.; Rotte, A.; Twomey, P.; Girish, S.; Wu, B. Personalized Cancer Vaccines: Clinical Landscape, Challenges, and Opportunities. Mol.Ther. 2021, 29, 555–570. [Google Scholar] [CrossRef]
- Xu, Z.; Fisher, D.E. mRNA melanoma vaccine revolution spurred by the COVID-19 pandemic. Front. Immunol. 2023, 14, 1155728. [Google Scholar] [CrossRef]


| NCT Number | Tumor Type/Target | Phase | Category | Status |
|---|---|---|---|---|
| NCT03190265 | Pancreatic Cancer | II | Tumor cell | Recruiting |
| NCT02451982 | Pancreatic Cancer | I/II | Tumor cell | Recruiting |
| NCT03767582 | Advanced PDAC | I/II | Tumor cell | Recruiting |
| NCT03376477 | Multiple Myeloma | II | Tumor cell | Recruiting |
| NCT03096093 | Neoplasms | I/II | Allogeneic cell | Recruiting |
| NCT03970746 | NSCLC | I/II | DC | Recruiting |
| NCT03059485 | AML | II | DC | Recruiting |
| NCT04523688 | Glioblastoma | II | DC | Not yet recruiting |
| NCT03136406 | Pancreatic Cancer/Mutant KRAS | I/II | Virus vector | Active, not recruiting |
| NCT03632941 | Breast Cancer/HER2 | II | Virus vector | Recruiting |
| NCT03547999 | Metastatic Colorectal Cancer/MVA-BN-CV301 | II | Virus vector | Active, not recruiting |
| NCT04747002 | Acute Myeloid Leukemia/DSP-7888 | II | Peptide | Recruiting |
| NCT04263051 | Advanced NSCLC/UCPVax | II | Peptide | Recruiting |
| NCT03149003 | Glioblastoma/DSP-7888 | III | Peptide | Recruiting |
| NCT04206254 | Liver Cancer/gp96 | II/III | Peptide | Not yet recruiting |
| NCT04274153 | Human Papilloma Virus/Gardasil9 | IV | Protein | Recruiting |
| NCT04090528 | Prostate Cancer, pTVG-HP, pTVG-AR | II | DNA | Recruiting |
| NCT03721978, | Cervical cancer/VGX-3100 | III | DNA | Recruiting |
| NCT04526899 | Melanoma Stage III-IV/NY-ESO-1, MAGE-A3, Tyrosinase, and TPTE | II | mRNA | Recruiting |
| NCT04163094 | Ovarian Cancer W-ova1 | I | mRNA | Recruiting |
| NCT03394937 | Resected melanoma (stages IIc, III, and IV)/CD40L, CD70, caTLR4, gp100, MAGE-A3, MAGE-C2, and PRAME | I | mRNA | Recruiting |
| NCT02410733 | Melanoma/1 NY-ESO-1, tyrosinase, MAGE-A3, and TPTE | I | mRNA | Recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katopodi, T.; Petanidis, S.; Grigoriadou, E.; Anestakis, D.; Charalampidis, C.; Chatziprodromidou, I.; Floros, G.; Eskitzis, P.; Zarogoulidis, P.; Koulouris, C.; et al. Immune Specific and Tumor-Dependent mRNA Vaccines for Cancer Immunotherapy: Reprogramming Clinical Translation into Tumor Editing Therapy. Pharmaceutics 2024, 16, 455. https://doi.org/10.3390/pharmaceutics16040455
Katopodi T, Petanidis S, Grigoriadou E, Anestakis D, Charalampidis C, Chatziprodromidou I, Floros G, Eskitzis P, Zarogoulidis P, Koulouris C, et al. Immune Specific and Tumor-Dependent mRNA Vaccines for Cancer Immunotherapy: Reprogramming Clinical Translation into Tumor Editing Therapy. Pharmaceutics. 2024; 16(4):455. https://doi.org/10.3390/pharmaceutics16040455
Chicago/Turabian StyleKatopodi, Theodora, Savvas Petanidis, Eirini Grigoriadou, Doxakis Anestakis, Charalampos Charalampidis, Ioanna Chatziprodromidou, George Floros, Panagiotis Eskitzis, Paul Zarogoulidis, Charilaos Koulouris, and et al. 2024. "Immune Specific and Tumor-Dependent mRNA Vaccines for Cancer Immunotherapy: Reprogramming Clinical Translation into Tumor Editing Therapy" Pharmaceutics 16, no. 4: 455. https://doi.org/10.3390/pharmaceutics16040455






