Genome Sequence and Characterisation of Peribacillus sp. Strain AS_2, a Bacterial Endophyte Isolated from Alectra sessiliflora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of the Bacterial Peribacillus sp. Strain AS_2
2.2. Genome Extraction, Library Preparation, and Sequencing
2.3. Genome De Novo Assembly and Annotation
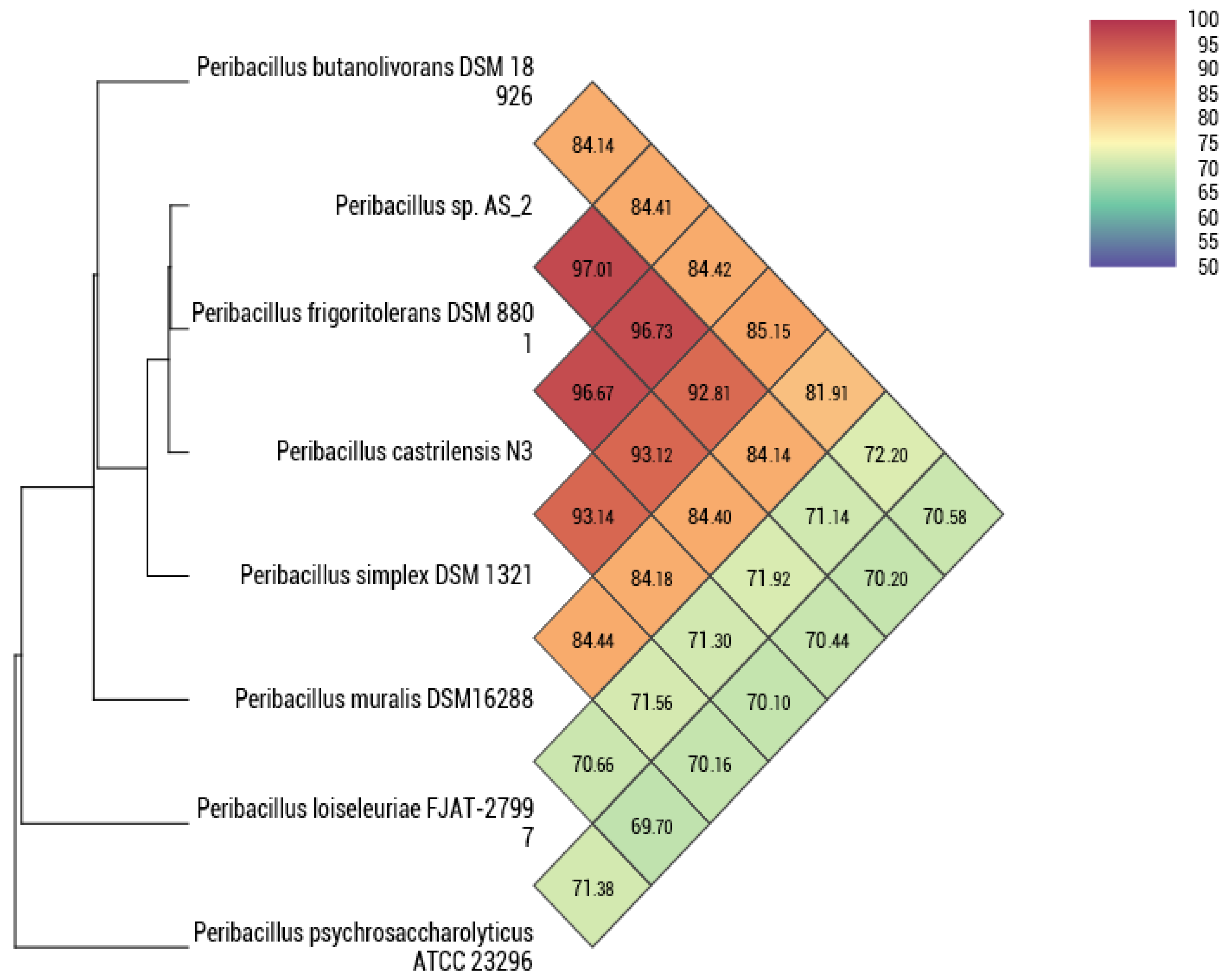
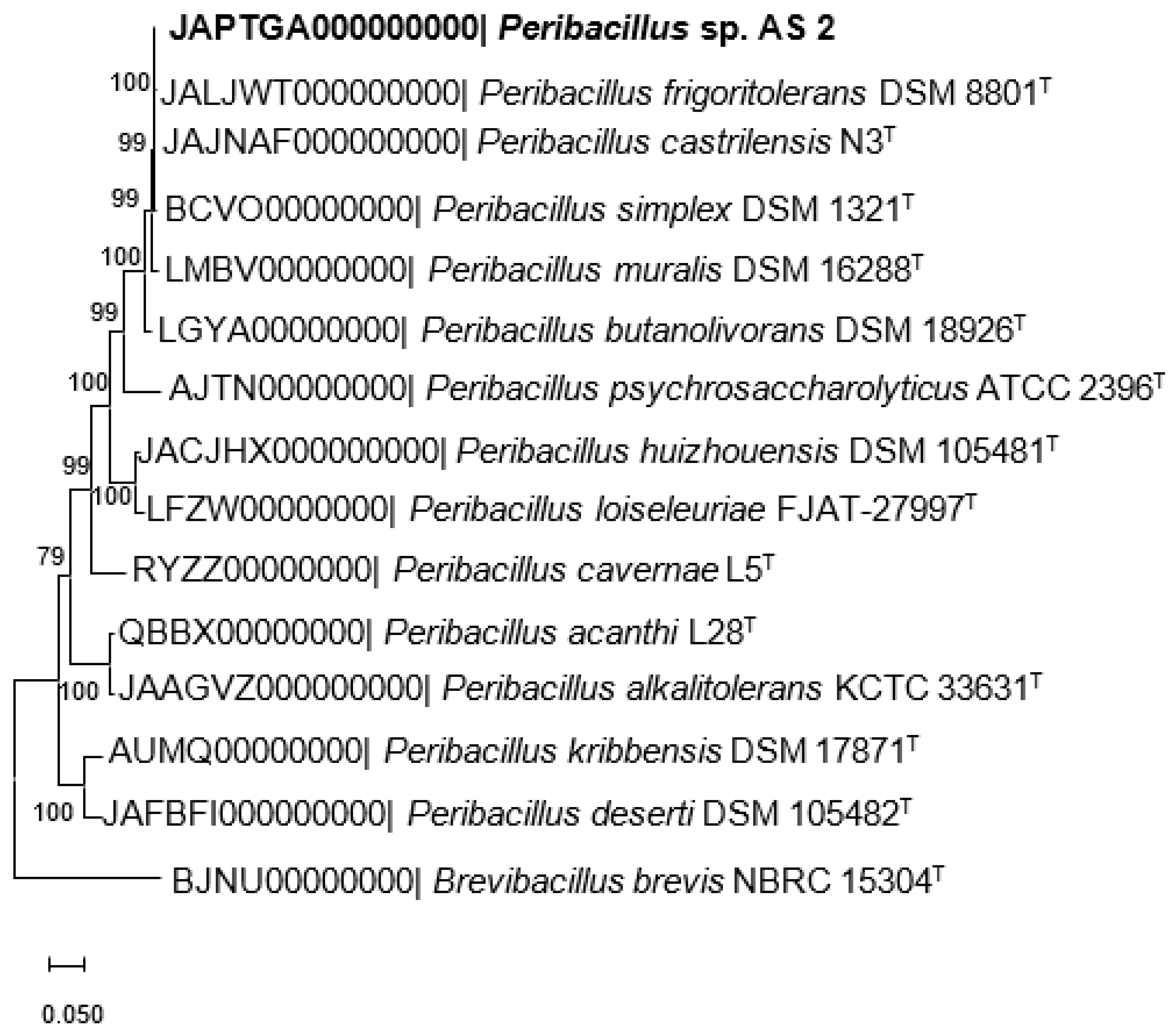
2.4. Phylogenome Analysis
2.5. Phenotypic Characterisation
2.6. Statistical Analysis
3. Results and Discussion
3.1. Genome Characteristics of Peribacillus sp. Strain AS_2
3.2. Functional Annotation
3.2.1. In Silico Analysis of Biosynthetic Gene Clusters
3.2.2. Antibiotic Resistance Genes
3.2.3. Genes Involved in Endophytic Lifestyle
3.2.4. Genes Involved in Biodegradation
3.3. Phenotypic Characterisation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, L.; Yang, C.; Wang, Y.; Ma, T.; Cai, F.; Wei, L.; Jin, M.; Osei, R.; Zhang, J.; Tang, M. Potential of an endophytic bacteria Bacillus amyloliquefaciens 3–5 as biocontrol agent against potato scab. Microb. Pathog. 2022, 163, 105382. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.L.; Bae, H. Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 2022, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J. Trichoderma parareesei favors the tolerance of rapeseed (Brassica napus L.) to salinity and drought due to a chorismate mutase. Agronomy 2020, 10, 118. [Google Scholar] [CrossRef]
- Sharma, S.; Dhar, M.K.; Kaul, S. Antagonistic, plant growth promoting and extracellular hydrolytic enzyme activity of fungal endophytes of Dioscorea bulbifera L. Biocatal. Agric. Biotechnol. 2023, 50, 102694. [Google Scholar] [CrossRef]
- Poveda, J.; Eugui, D.; Abril-Urías, P.; Velasco, P. Endophytic fungi as direct plant growth promoters for sustainable agricultural production. Symbiosis 2021, 85, 1–9. [Google Scholar] [CrossRef]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; De Vos, P.; Genus, I. Bacillus. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2009; Volume 3, pp. 21–128. [Google Scholar]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar]
- Patel, S.; Gupta, R.S. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar]
- Narsing Rao, M.P.; Dhulappa, A.; Banerjee, A.; Thamchaipenet, A. Transfer of Bacillus tepidiphilus Narsing Rao et al. 2021 to the genus Peribacillus as Peribacillus tepidiphilus comb. nov. Arch. Microbiol. 2022, 204, 545. [Google Scholar] [CrossRef]
- Ma, K.; Yin, Q.; Chen, L.; Lai, Q.; Xu, Y. Bacillus acanthi sp. nov., isolated from the rhizosphere soil of a mangrove plant Acanthus ilicifolius. Int. J. Syst. Evol. Microbiol. 2018, 68, 3047–3051. [Google Scholar] [CrossRef]
- Jiang, L.; Jung, W.Y.; Li, Z.; Lee, M.-K.; Park, S.-H.; Kang, S.W.; Lee, J.-S.; Jung, H.; Hur, T.-Y.; Kim, H.B.; et al. Peribacillus faecalis sp. nov., a moderately halophilic bacterium isolated from the faeces of a cow. Int. J. Syst. Evol. Microbiol. 2021, 71, 004721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Chen, W.F.; Li, M.; Sui, X.H.; Liu, H.C.; Zhang, X.X.; Chen, W.X. Bacillus endoradicis sp. nov., an endophytic bacterium isolated from soybean root. Int. J. Syst. Evol. Microbiol. 2012, 62, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Busse, H.J.; McInroy, J.A.; Glaeser, S.P. Bacillus gossypii sp. nov., isolated from the stem of Gossypium hirsutum. Int. J. Syst. Evol. Microbiol. 2015, 65, 4163–4168. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Reina, J.C.; Sampedro, I.; Llamas, I.; Martínez-Checa, F. Peribacillus castrilensis sp. nov.: A Plant-Growth-Promoting and Biocontrol Species Isolated From a River Otter in Castril, Granada, Southern Spain. Front. Plant Sci. 2022, 13, 896728. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Kim, S.; An, J.H.; Sang, M.K.; Weon, H.Y. Complete genome sequence of Peribacillus butanolivorans KJ40, a soil bacterium alleviating drought stress in plants. Korean J. Microbiol. 2020, 56, 407–409. [Google Scholar]
- Sato, S.; Ichiyanagi, N.; Sugiyama, K.; Aburai, N.; Fujii, K. Production of polyglutamic acid-like mucilage protein by Peribacillus simplex strain 8h. Folia. Microbiol. 2023, 68, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, D.; Srivastava, A.; Srivastava, P.C.; Sharma, A. Evaluation of three novel soil bacterial strains for efficient biodegradation of persistent boscalid fungicide: Kinetics and identification of microbial biodegradation intermediates. Environ. Pollut. 2023, 316, 120484. [Google Scholar] [CrossRef] [PubMed]
- Maela, M.P.; van der Walt, H.; Serepa-Dlamini, M.H. The antibacterial, antitumor activities, and bioactive constituents’ identification of ALECTRA sessiliflora bacterial endophytes. Front. Microbiol. 2022, 13, 870821. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. J. Nucleic Acids 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 12 August 2023).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. J. Bioinform. 2013, 15, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. J. Nucleic Acids 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Abby, S.S.; Néron, B.; Ménager, H.; Touchon, M.; Rocha, E.P. MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS ONE 2014, 9, e110726. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon, F.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.; Webster, A.L.; Cao, M.P.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; Berge, O.; Bishop, A.H.; Busse, H.J.; De Vos, P.; Fritze, D.; Heyndrickx, M.; Kampfer, P.; Rabinovitch, L.; Salkinoja-Salonen, M.S.; et al. Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Montecillo, J.A.; Bae, H. Reclassification of Brevibacterium frigoritolerans as Peribacillus frigoritolerans comb. nov. based on phylogenomics and multiple molecular synapomorphies. Int. J. Syst. Evol. 2022, 72, 005389. [Google Scholar] [CrossRef]
- Liu, G.H.; Liu, B.; Wang, J.P.; Che, J.M.; Li, P.F. Reclassification of Brevibacterium frigoritolerans DSM 8801 T as Bacillus frigoritolerans comb. nov. Based on Genome Analysis. Curr. Microbiol. 2020, 77, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, K.; Kube, M.; Becker, R.; Schneck, V.; Ulrich, A. Genomic analysis of the endophytic Stenotrophomonas strain 169 reveals features related to plant-growth promotion and stress tolerance. Front. Microbiol. 2021, 12, 687463. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E.; Tyler, H.L.; DeBoy, R.T.; Daugherty, S.; Ren, Q.; Badger, J.H.; Durkin, A.S.; Huot, H.; Shrivastava, S.; Kothari, S.; et al. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 2008, 4, e1000141. [Google Scholar] [CrossRef]
- Guo, F.B.; Xiong, L.; Zhang, K.Y.; Dong, C.; Zhang, F.Z.; Woo, P.C. Identification and analysis of genomic islands in Burkholderia cenocepacia AU 1054 with emphasis on pathogenicity islands. BMC Microbiol. 2017, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Duan, J.; Charles, T.C.; Glick, B.R. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J. Theor. Biol. 2014, 343, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, K.; Miyazaki, K. Revisiting bacterial phylogeny: Natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob. Genet. Elem. 2013, 3, e24210. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wang, Q.; Li, M.; Wang, S.; Ye, K.; Dai, W.; Huang, J. Culturable bacterial endophytes of Aconitum carmichaelii Debx. were diverse in phylogeny, plant growth promotion, and antifungal potential. Front. Microbiol. 2023, 14, 1192932. [Google Scholar] [CrossRef]
- Verma, V.C.; Gange, A.C. (Eds.) Advances in Endophytic Research; Springer Science & Business Media: New Delhi, India, 2013. [Google Scholar]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech 2017, 7, 315. [Google Scholar] [CrossRef]
- Akter, Y.; Barua, R.; Nasir Uddin, M.; Muhammad Sanaullah, A.F.; Marzan, L.W. Bioactive potentiality of secondary metabolites from endophytic bacteria against SARS-COV-2: An in-silico approach. PLoS ONE 2022, 17, e0269962. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Sun, Z.; Li, Y.; He, H.; Zhang, Y.; Yang, X.; Wang, D.; Dong, B.; Zhou, H.; et al. Whole-genome analysis revealed the growth-promoting mechanism of endophytic bacterial strain Q2H1 in potato plants. Front. Microbiol. 2022, 13, 1035901. [Google Scholar] [CrossRef]
- Montecillo, J.A.; Bae, H. In silico analysis of koranimine, a cyclic imine compound from Peribacillus frigoritolerans reveals potential nematicidal activity. Sci. Rep. 2022, 12, 18883. [Google Scholar] [CrossRef]
- Evans, B.S.; Ntai, I.; Chen, Y.; Robinson, S.J.; Kelleher, N.L. Proteomics-based discovery of koranimine, a cyclic imine natural product. J. Am. Chem. Soc. 2011, 133, 7316–7319. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Wolk, C.P.; Kuritz, T.; Sasamoto, S.; Watanabe, A.; Iriguchi, M.; Ishikawa, A.; Kawashima, K.; Kimura, T.; et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 (supplement). DNA Res. 2001, 8, 227–253. [Google Scholar] [CrossRef]
- Fresia, P.; Antelo, V.; Salazar, C.; Giménez, M.; D’Alessandro, B.; Afshinnekoo, E.; Mason, C.; Gonnet, G.H.; Iraola, G. Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters. Microbiome 2019, 7, 35. [Google Scholar] [CrossRef]
- Zhao, C.X.; Su, X.X.; Xu, M.R.; An, X.L.; Su, J.Q. Uncovering the diversity and contents of gene cassettes in class 1 integrons from the endophytes of raw vegetables. Ecotoxicol. Environ. Saf. 2022, 247, 114282. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Millan, A.S. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018, 26, 978–985. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 167, 76. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.S.; Dallman, T.J.; Field, N.; Childs, T.; Mitchell, H.; Day, M.; Weill, F.X.; Lefèvre, S.; Tourdjman, M.; Hughes, G.; et al. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat. Commun. 2018, 9, 1462. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Nosrati, M.; Dey, D.; Mehrani, A.; Strassler, S.E.; Zelinskaya, N.; Hoffer, E.D.; Stagg, S.M.; Dunham, C.M.; Conn, G.L. Functionally critical residues in the aminoglycoside resistance-associated methyltransferase RmtC play distinct roles in 30S substrate recognition. J. Biol. Chem. 2019, 294, 17642–17653. [Google Scholar] [CrossRef]
- Finken, M.; Kirschner, P.; Meier, A.; Wrede, A.; Böttger, E.C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Biol. 1993, 9, 1239–1246. [Google Scholar] [CrossRef]
- Samant, S.; Hsu, F.F.; Neyfakh, A.A.; Lee, H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 2009, 191, 1311–1319. [Google Scholar] [CrossRef]
- Jagielski, T.; Bakuła, Z.; Brzostek, A.; Minias, A.; Stachowiak, R.; Kalita, J.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Żaczek, A.; Vasiliauskiene, E.; et al. Characterization of mutations conferring resistance to rifampin in Mycobacterium tuberculosis clinical strains. Antimicrob. Agents Chemother. 2018, 62, e01093-18. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Lang, M.; Schneider, P.; Tosch, W.; Scartazzini, R.; Zak, O. The Penems, a New Class of β-Lactam Antibiotics 7. Synthesis and Antimicrobial Activity of 2-Heterocyclylmercaptoalkyl Derivatives. J. Antibiot. 1986, 39, 525–534. [Google Scholar] [CrossRef]
- Shastry, R.P.; Welch, M.; Rai, V.R.; Ghate, S.D.; Sandeep, K.; Rekha, P.D. The whole-genome sequence analysis of Enterobacter cloacae strain Ghats1: Insights into endophytic lifestyle-associated genomic adaptations. Arch. Microbiol. 2020, 202, 1571–1579. [Google Scholar] [CrossRef]
- Deng, W.; Li, C.; Xie, J. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell. Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef]
- Pinski, A.; Betekhtin, A.; Hupert-Kocurek, K.; Mur, L.A.; Hasterok, R. Defining the genetic basis of plant–endophytic bacteria interactions. Int. J. Mol. Sci. 2019, 20, 1947. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Park, S.C.; Kwak, Y.M.; Song, W.S.; Hong, M.; Yoon, S.I. Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR. Nucleic Acids Res. 2017, 45, 13080–13093. [Google Scholar] [CrossRef]
- Ko, M.; Park, C. H-NS-Dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 2000, 182, 4670–4672. [Google Scholar] [CrossRef]
- Diale, M.O.; Kayitesi, E.; Serepa-Dlamini, M.H. Genome in silico and in vitro analysis of the probiotic properties of a bacterial endophyte, Bacillus paranthracis strain mhsd3. Front. Genet. 2021, 12, 672149. [Google Scholar] [CrossRef]
- O’Neill, J. Review on antimicrobial resistance: Tackling drug-resistant infections globally: Final report and recommendations. In Review on Antimicrobial Resistance: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Wellcome Trust: London, UK, 2016. [Google Scholar]
- Yamaguchi, M.; Goto, K.; Hirose, Y.; Yamaguchi, Y.; Sumitomo, T.; Nakata, M.; Nakano, K.; Kawabata, S. Identification of evolutionarily conserved virulence factor by selective pressure analysis of Streptococcus pneumoniae. Commun. Biol. 2019, 2, 96. [Google Scholar] [CrossRef]
- Lòpez-Fernàndez, S.; Sonego, P.; Moretto, M.; Pancher, M.; Engelen, K.; Pertot, I.; Campisano, A. Whole-genome comparative analysis of virulence genes unveils similarities and differences between endophytes and other symbiotic bacteria. Front. Microbiol. 2015, 6, 419. [Google Scholar]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef]
- Fernandez, R.C.; Weiss, A.A. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun. 1994, 62, 4727–4738. [Google Scholar] [CrossRef]
- Kendrew, S.G.; Federici, L.; Savino, C.; Miele, A.; Marsh, E.N.; Vallone, B. Crystallization and preliminary X-ray diffraction studies of a monooxygenase from Streptomyces coelicolor A3 (2) involved in the biosynthesis of the polyketide actinorhodin. Acta Crystallogr. D Struct. Biol. 2000, 56, 481–483. [Google Scholar] [CrossRef]
- Ye, J.; Dickens, M.L.; Plater, R.; Li, Y.; Lawrence, J.; Strohl, W.R. Isolation and sequence analysis of polyketide synthase genes from the daunomycin-producing Streptomyces sp. strain C5. J. Bacteriol. 1994, 176, 6270–6280. [Google Scholar] [CrossRef]
- Grocholski, T.; Oja, T.; Humphrey, L.; Mäntsälä, P.; Niemi, J.; Metsä-Ketelä, M. Characterization of the two-component monooxygenase system AlnT/AlnH reveals early timing of quinone formation in alnumycin biosynthesis. J. Bacteriol. 2012, 194, 2829–2836. [Google Scholar] [CrossRef]
- Yoon, E.J.; Jeong, S.H. Class D β-lactamases. J. Antimicrob. Chemother. 2021, 76, 836–864. [Google Scholar] [CrossRef]
- Abubakar, I.I.; Tillmann, T.; Banerjee, A. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar]
- Troeger, C.; Forouzanfar, M.; Rao, P.C.; Khalil, I.; Brown, A.; Reiner, R.C.; Fullman, N.; Thompson, R.L.; Abajobir, A.; Ahmed, M.; et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef]
- Hesham, A.E.L.; Mawad, A.M.; Mostafa, Y.M.; Shoreit, A. Biodegradation ability and catabolic genes of petroleum-degrading Sphingomonas koreensis strain ASU-06 isolated from Egyptian oily soil. BioMed Res. Int. 2014, 2014, 127674. [Google Scholar] [CrossRef]
- Karaś, M.A.; Wdowiak-Wróbel, S.; Sokołowski, W. Selection of endophytic strains for enhanced bacteria-assisted phytoremediation of organic pollutants posing a public health hazard. Int. J. Mol. Sci. 2021, 22, 9557. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Balgobind, A.; Pillay, B. Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 2013, 14, 10197–10228. [Google Scholar] [CrossRef]
- Santero, E.; Díaz, E. Special Issue: Genetics of biodegradation and bioremediation. Genes 2020, 11, 441. [Google Scholar] [CrossRef] [PubMed]
- Balogh-Hergovich, É.; Kaizer, J.; Speier, G. Chemical models relevant to nitroalkane dioxygenase. Comptes Rendus Chim. 2007, 10, 355–365. [Google Scholar] [CrossRef]
- Torres-Guzman, J.C.; Padilla-Guerrero, I.E.; Cervantes-Quintero, K.Y.; Martinez-Vazquez, A.; Ibarra-Guzman, M.; Gonzalez-Hernandez, G.A. Peculiarities of nitronate monooxygenases and perspectives for in vivo and in vitro applications. Appl. Microbiol. Biotechnol. 2021, 105, 8019–8032. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, Z.; Hou, H.; Li, L.; Zhang, J.; Yang, H.; Dong, Y.; Tan, H. Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes. Biochem. Biophys. Res. Commun. 2011, 405, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Shin, B.; Park, W. Protective role of bacterial alkanesulfonate monooxygenase under oxidative stress. Appl. Microbiol. Biotechnol. 2020, 86, e00692-20. [Google Scholar] [CrossRef] [PubMed]
- Haarstad, K.; Bavor, H.J.; Mæhlum, T. Organic and metallic pollutants in water treatment and natural wetlands: A review. Water Sci. Technol. 2012, 65, 76–99. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, K.; Malik, A. Screening of polycyclic aromatic hydrocarbon degrading bacterial isolates from oil refinery wastewater and detection of conjugative plasmids in polycyclic aromatic hydrocarbon tolerant and multi-metal resistant bacteria. Heliyon 2019, 5, e02742. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Xiong, W.; Qiu, H.; Peng, W.; Deng, Z.; Lin, S.; Liang, R. Characterization of the phenanthrene-degrading Sphingobium yanoikuyae SJTF8 in heavy metal co-existing liquid medium and analysis of its metabolic pathway. Microorganisms 2020, 8, 946. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Enquahone, S.; van Marle, G.; Simachew, A. Plant growth-promoting characteristics of halotolerant endophytic bacteria isolated from Sporobolus specatus (Vahr) Kunth and Cyperus laevigatus L. of Ethiopian rift valley lakes. Arch. Microbiol. 2022, 204, 403. [Google Scholar] [CrossRef]
- Heyrman, J.; Logan, N.A.; Rodríguez-Díaz, M.; Scheldeman, P.; Lebbe, L.; Swings, J.; Heyndrickx, M.; De Vos, P. Study of mural painting isolates, leading to the transfer of ‘Bacillus maroccanus’ and ‘Bacillus carotarum’to Bacillus simplex, emended description of Bacillus simplex, re-examination of the strains previously attributed to ‘Bacillus macroides’ and description of Bacillus muralis sp. nov. J. Syst. Evol. Microbiol. 2005, 55, 119–131. [Google Scholar]
- Kuisiene, N.; Raugalas, J.; Sproer, C.; Kroppenstedt, R.M.; Chitavichius, D. Bacillus butanolivorans sp. nov., a species with industrial application for the remediation of n-butanol. J. Syst. Evol. Microbiol. 2008, 58, 505–509. [Google Scholar] [CrossRef] [PubMed]


| Genome Characteristic | Value |
|---|---|
| Genome size (bp) | 5,482,853 |
| G + C content (%) | 40.5 |
| Total number of genes | 5290 |
| Protein coding genes | 5072 |
| Number of RNAs | 88 |
| rRNA genes | 1, 1, 1 (5S, 16S, 23S) |
| tRNA genes | 79 |
| ncRNAs | 6 |
| Pseudogenes | 130 |
| CRISPR repeats | 4 |
| Cas cluster | 1 |
| From (bp) | To (bp) | Type | Production | Similarity (%) | Reference |
|---|---|---|---|---|---|
| 236,408 | 296,726 | NRPs | Koranimine | 87 | Bacillus sp. NK2003 [56] |
| 140,936 | 165,105 | Beta-lactone | Fengycin | 46 | Bacillus velezensis FZB42 [57] |
| 50,380 | 65,892 | NI-siderophore | Schizokinen | 60 | Nostoc sp. PCC 7120 [58] |
| 380,256 | 402,151 | Terpene | - | - | |
| 389,596 | 430,684 | T3PKS | - | - | |
| 87,936 | 132,003 | NRPS | - | - | |
| 8889 | 32,424 | LAP | - | - | |
| 7431 | 28,249 | Terpene | - | - |
| Characteristic | 1 | 2+ | 3+ | 4+ | 5+ |
|---|---|---|---|---|---|
| Anaerobic growth | + | - | v | w | - |
| Growth at 4 °C | - | + | - | - | + |
| Growth at 45 °C | + | - | - | - | + |
| NaCl (w/v, %) | 0–7 | 0.5–7.5 | <5 | <7 | 0.5–5 |
| Optimum NaCl (w/v, %) | 1 | 0 | 0 | 0 | 1 |
| pH range | 3–12 | 5–10 | 6–9 | 6–9 | 6–9 |
| Optimum pH | 7 | 7 | 8 | 7 | 7 |
| Oxidase | + | - | - | + | + |
| API 20E: | |||||
| ONPG | - | - | - | + | - |
| Arginine dihydrolase | - | - | - | - | - |
| Citrate utilisation | - | - | - | - | - |
| Voges– Proskauer | + | - | - | - | - |
| Urease | - | - | - | - | - |
| Tryptophan deaminase | - | - | - | - | - |
| Indole production | - | - | - | - | - |
| Gelatine hydrolysis | - | - | - | - | - |
| API 50: | |||||
| L-arabinose | + | + | - | + | - |
| Ribose | + | - | - | + | - |
| Glucose | + | + | w | + | - |
| Fructose | + | + | w | + | - |
| Mannitol | + | - | - | + | - |
| N-acetylglucosamine | + | - | w | + | w |
| Salicin | + | - | w/v | + | w/v |
| Saccharose | + | - | - | - | - |
| Trehalose | + | - | w | + | w |
| Inulin | + | - | w | - | w |
| Raffinose | + | + | - | + | - |
| Hydrolysis of: | |||||
| Starch | - | - | + | + | - |
| Casein | + | + | v | v | - |
| DNA G + C content (mol %) | 40.45 | 40.6 | 39.9 | 41.2 | 37.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maela, M.P.; Serepa-Dlamini, M.H. Genome Sequence and Characterisation of Peribacillus sp. Strain AS_2, a Bacterial Endophyte Isolated from Alectra sessiliflora. Microbiol. Res. 2024, 15, 50-65. https://doi.org/10.3390/microbiolres15010004
Maela MP, Serepa-Dlamini MH. Genome Sequence and Characterisation of Peribacillus sp. Strain AS_2, a Bacterial Endophyte Isolated from Alectra sessiliflora. Microbiology Research. 2024; 15(1):50-65. https://doi.org/10.3390/microbiolres15010004
Chicago/Turabian StyleMaela, Mehabo Penistacia, and Mahloro Hope Serepa-Dlamini. 2024. "Genome Sequence and Characterisation of Peribacillus sp. Strain AS_2, a Bacterial Endophyte Isolated from Alectra sessiliflora" Microbiology Research 15, no. 1: 50-65. https://doi.org/10.3390/microbiolres15010004







