Abstract
Mcm1 is an essential Q/N-rich transcription factor. Q/N-rich proteins interact with each other, and many affect the [PSI+] prion formed by the translation termination factor Sup35 (eRF3). We found that transient MCM1 overexpression increased nonsense suppression in [PSI+] strains and SUP35 transcription. As we had discovered similar effects of another Q/N-rich transcription factor, Sfp1, here we focus on the roles of Mcm1 and Sfp1 in SUP35 expression, as well as on the effects of Sfp1 on the expression of the gene encoding another release factor, Sup45 (eRF1). Mutations in the SUP35 promoter showed that none of the potential Mcm1 binding sites affected the Sup35 protein level or nonsense suppression, even during MCM1 overexpression. Mcm1 itself neither formed aggregates in vivo nor affected Sup35 aggregation. In contrast, a mutation in the Sfp1-binding site decreased Sup35 production and [PSI+] toxicity of excess Sfp1. Mutation of the Sfp1 binding site in the SUP45 promoter lowered SUP45 expression and increased nonsense suppression even more drastically. Our data indicate that the mechanisms of Mcm1 and Sfp1 action differ. While Mcm1 seems unlikely to directly regulate SUP35 expression, Sfp1 appears to act through its binding sites and to directly activate SUP35 expression, which in turn may influence the [PSI+] prion phenotype and toxicity.
1. Introduction
Amyloid prions in baker’s yeast (Saccharomyces cerevisiae) act as heritable cytosolic factors, as the newly synthesized proteins rapidly acquire amyloid conformation, which is then transmitted to the daughter cells and thereby persists in the cell progeny. [PSI+] is a prion formed by self-perpetuating amyloid of yeast essential release factor Sup35 (eRF3). Presence of [PSI+] leads to almost complete inactivation of Sup35 in an aggregated form, resulting in shortage of release factor and inefficient termination of translation. This in turn causes more frequent stop codon readthrough, and may lead to suppression of nonsense mutations (or nonsense suppression), as reviewed in []. Two release factors, Sup35 and Sup45 (eRF1), are involved in the termination of translation. Nonsense suppression may also result from mutations in either of these factors or from their downregulation (reviewed in [,,]).
Most known yeast prion proteins contain domains that are enriched in asparagine (N) or glutamine (Q) residues, which are necessary for prion maintenance (reviewed in [,]). In the case of the Sup35 protein, its N-terminal domain is Q/N-rich and prionogenic, its middle (M) domain is half positively and half negatively charged, aiding in its possible pH-sensor function during phase separation into biomolecular condensates [], and the C-terminal domain is globular and essential for the eRF3 release factor function [,]. Yeast amyloid prions are known to interact with each other; for example, [PIN+], a prion form of another Q/N-rich protein, Rnq1, is necessary for the de novo appearance of [PSI+], i.e., it acts as a [PSI+]-inducing (Pin+) factor []. While [PIN+] has no detectable phenotype, its presence along with another prion [SWI+] formed by the transcription factor Swi1 results in the [NSI+] factor, which manifests in a [PSI+]-like nonsense suppressor phenotype caused by SUP45 downregulation [,]. These interactions are not limited to only prion-forming proteins; various other Q/N-rich proteins have been shown to act as Pin+-factors [] as well as to form nonheritable aggregates [,] that can indirectly affect prion properties. Notably, transcription factors are often found among Q/N-rich proteins, with at least four of them forming bona fide prions: Ure2, Swi1, Mot3, and Cyc8 (reviewed in [,]).
We previously investigated the possible influence of two Q/N-rich transcription factors, Mcm1 and Sfp1, on [PSI+] properties. Both were discovered by screening using a synthetic lethality test, which helped to identify factors that affect the lethality of [PSI+] in the presence of sup45 mutation by influencing nonsense suppression [,,]. Here, we attempted to find out whether the effects of Mcm1 and Sfp1 on [PSI+] are due to their function as transcription regulators.
2. Materials and Methods
2.1. Plasmids
All of the plasmids used in this work are listed in Supplementary Table S1. Plasmids pRS316 and pRS426 have been described before [,]. Plasmids pRS426-SFP1 [] and pU-MCM1 [] were kindly provided by Tatiana M. Rogoza and Anton A. Nizhnikov, respectively. pUGC-MCM1-GFP is a pUG35-based vector obtained through a series of intermediate vectors and contains the MCM1 ORF with upstream 50 bp sequence, flanked by BglII and SacII restriction sites (the PsuI-SacII fragment of the pGPD-f1-MCM1-YFP plasmid []) under control of the CUP1 promoter originally derived from pRS316CG []. Plasmid pYX242-Nab2NLS-2mCherry [] was a kind gift from Simon Alberti. The plasmids with mutations and deletions in the SUP35 and SUP45 promoters are based on pRSU1 [] and pRS315-SUP45 [], respectively. They were constructed using site-directed mutagenesis as described previously in []. The primers used for site-directed mutagenesis are listed in Supplementary Table S2. The ΔMcm1-2* variant, which has an insertion of an additional C in position −282 as well as a one-nucleotide deletion in the Abf1 binding site (mutAbf1 variant), was obtained as a PCR-induced mutation during site-directed mutagenesis. All the obtained vectors were verified by Sanger sequencing.
2.2. Strains
The yeast strains used in this work are listed in Supplementary Table S3. The strains used for MCM1 overexpression studies were isogenic to 74-D694 [,,,,]. All other strains, which were used for plasmid shuffling experiments, were isogenic to GT81 [,,,,,]. Strains with LEU2 plasmids with mutant promoter variants were selected via plasmid shuffling using 5-FOA medium as described previously [].
Yeasts were grown at 30 °C in standard liquid and solid media using conventional methods [,]. 1/4YEPD medium was used for the color phenotype detection []. To check for the presence of the [PSI+] prion, 1/4YEPD medium supplemented with 4 mM GuHCl was used. For the induction of the CUP1 promoter, CuSO4 was routinely added to the medium at a final concentration of 50 M unless indicated otherwise. Yeast transformations were performed using a standard protocol [].
2.3. qPCR
Cells were grown in liquid cultures to the mid-log phase. In the case of CUP1 promoter induction, the medium was supplemented with CuSO4 at a final concentration of 150 M. RNA extraction, cDNA synthesis, and qPCR reactions were performed as described previously []. The primers used for the qPCR are listed in Supplementary Table S2. ACT1 was used as a reference. Relative units of expression were calculated as 2−Ct [].
2.4. Protein Analysis
The alkaline lysis protocol [] was used for protein extraction for subsequent SDS-PAGE and Western blot analysis []. SDD-AGE and capillary transfer were performed as described [,]. The antibodies SE4290 [], SE-45-2 [], Anti-GFP (Abcam, Cambridge, UK, #ab290), ADH1A (LsBio, Lynnwood, WA, USA, #LS-C68862), and Anti--Tubulin (Sigma-Aldrich, St. Louis, MO, USA, #T6074) were used to detect Sup35, Sup45, GFP, Adh1, and Tub1, respectively. ECL Select Western Blotting Detection Reagent (Cytiva, Marlborough, MA, USA) was used for antibody detection. Images were acquired using GeneGnome (Syngene, Bangalore, India).
2.5. Fluorescence Microscopy
Cells were grown in liquid media until reaching OD600 = 0.2–0.3. CuSO4 was then added to a final concentration of 50 M for the CUP1 promoter induction. Cells were grown for an additional 3–4 h and then visualized using Zeiss Axioscope A1 equipped with a Zeiss AxioCam 506 Color camera. Images were acquired using Zeiss Zen software, version 3.9.
2.6. Bioinformatic Analysis
Searches for potential TFBSs in the SUP35 and SUP45 promoters was carried out using the oPOSSUM-3 online tool [] (http://cisreg.ca/software/, accessed on 27 March 2024). A similarity threshold of 75% was used. Mutant promoter variants were checked for the absence of pre-existing TFBS in the results, i.e., the similarity of all mutant sites to their respective TFBS profiles should be less than 75%. Transcription factor binding site profiles were taken from the JASPAR 2022 database [] (https://jaspar2022.genereg.net, accessed on 27 March 2024). Statistical analysis was performed in R v.4.3 (R Core Team, 2023). Boxplots were constructed using the ggplot2 package [].
3. Results
3.1. Transient Overexpression of MCM1 Enhances Nonsense Suppression in [PSI+] Strains
In our previous studies using constructs for the constitutive expression of the MCM1, we did not observe any effect on nonsense suppression; however, overexpression of the MCM1 gene controlled by copper-inducible CUP1 promoter appeared to enhance the synthetic lethality of [PSI+] with sup45 mutations in the test (Supplementary Figure S1). As the enhanced lethality in the test could reflect increased nonsense suppression, we checked whether transient Mcm1 overexpression affected the suppressor phenotype in [PSI+] strains. Indeed, various [PSI+] strains demonstrated enhanced suppression (Figure 1A). The C-terminally GFP-tagged variant of Mcm1 (Mcm1-GFP) showed the same effects (Figure 1A). Previously, we observed similar effects on the part of another Q/N-rich transcription factor Sfp1, which was shown to influence both transcription of the release factor genes and Sup35 aggregation []. Thus, we checked whether Mcm1 also affected these processes.
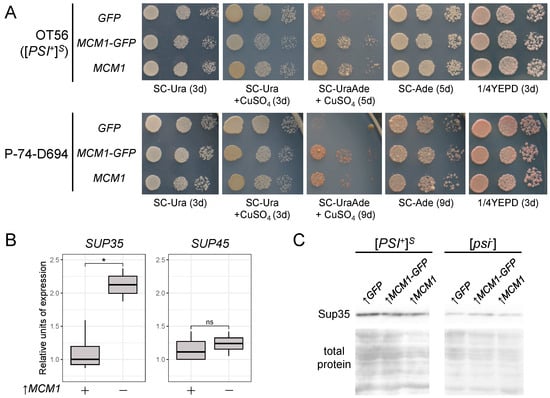
Figure 1.
MCM1 overexpression enhances nonsense suppression in [PSI+] strains. (A) The OT56 and P-74-D694 strains were transformed with pRS316CG (GFP), pUGC-MCM1-GFP (MCM1-GFP), or pU-MCM1 (MCM1). The phenotypes of the resulting clones were analyzed by plating cells onto various media to assess growth (SC-Ura, SC-Ura+CuSO4) and nonsense suppression (SC-UraAde+CuSO4, SC-Ade, 1/4YEPD). Shown are tenfold serial dilutions of the representative clones. (B) qPCR analysis of SUP35 and SUP45 mRNA levels in the OT56 ([PSI+]S) clones bearing the pU-MCM1 plasmid. The relative expression from copper-induced cultures is compared to no-induction samples. *, p < 0.05 in Wilcoxon Mann–Whitney test; ns, not significant (p > 0.05). (C) Analysis of the total Sup35 protein levels in OT56 ([PSI+]S) and 74-D694 ([psi−]) strains transformed with the plasmids from (A) using SDS-PAGE and Western blotting with anti-Sup35 antibodies. Coomassie R-250 staining was used to visualize the total proteins.
First, using qPCR, we assessed changes in SUP35 and SUP45 mRNA levels under transient overexpression of MCM1. A significant increase in SUP35 but not SUP45 mRNA levels was detected (Figure 1B), suggesting that MCM1 might be involved in the control of SUP35 transcription. However, no changes in Sup35 protein levels were observed (Figure 1C). These results were dissimilar to those obtained when studying Sfp1 overexpression, as the latter was shown to increase both SUP35 and SUP45 mRNA levels even though only Sup35 protein levels were visibly elevated []. Nevertheless, our results suggest that both Mcm1 and Sfp1 might be involved in regulating transcription of the release factor genes.
3.2. Search for the Potential Mcm1 and Sfp1 Binding Sites in the SUP35 and SUP45 Promoter Regions
We next attempted to determine whether Sfp1 and Mcm1 could directly regulate transcription of the SUP35 and SUP45 genes. We performed a bioinformatic analysis of the SUP35 and SUP45 promoters in order to find possible transcription factor binding sites (TFBSs) using the oPOSSUM 3.0 tool. We found four potential Mcm1 TFBSs in the SUP35 upstream region and six in the SUP45 upstream region. Three and two potential Sfp1 TFBSs were also found in the upstream regions of SUP35 and SUP45, respectively (Table 1). We designed mutations and deletions in the potential Mcm1 and Sfp1 TFBSs. Because no influence of the Mcm1 on SUP45 mRNA was observed, we chose only Mcm1 TFBSs in the SUP35 promoter (Figure 2A). We designed complete deletions of two such TFBSs and mutations in one. The latter site overlaps with TFBS of another transcription factor, Spt2, which is known to physically interact with the SUP35 promoter []. We changed the sequence in such a way that Spt2 TFBS would remain while the potential Mcm1 TFBS was lost (Figure 2A,B). We also designed mutations in the two most probable Sfp1 TFBSs, one in the SUP45, and one in the SUP35 promoter. The mutation in the SUP45 promoter was designated ‘flipSfp1’, as part of the sequence was flipped (Figure 2C).

Table 1.
Bioinformatic analysis of potential Mcm1 and Sfp1 binding sites in the SUP35 and SUP45 promoters Putative transcription factor binding sites (TFBS) of Sfp1 and Mcm1 in the promoter regions of SUP35 and SUP45 were identified by oPOSSUM3.0 single-site analysis. The %Score value indicates the similarity of the sequence to the TFBS profile. The optimal sites selected for further analysis are highlighted in bold.
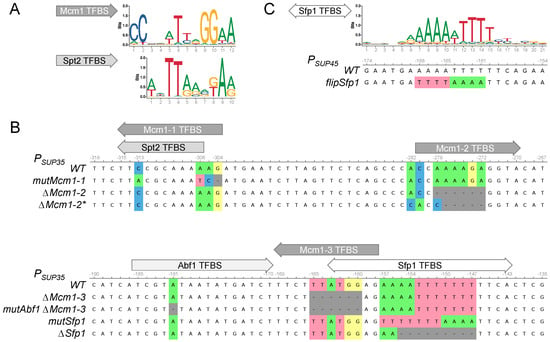
Figure 2.
Design of mutations and deletions in the potential Mcm1 and Sfp1 TFBS. (A) Profiles for the TFBSs of Mcm1 (MA0331.1) and Spt2 (MA0387.1) according to the JASPAR2022 database. (B) Locations of the predicted TFBSs in the SUP35 promoter and alignments of the deletions and mutations used in this work. (C) Profile for the Sfp1 TFBS (MA0378.1) and alignment of the SUP45 promoter region predicted to contain this site.
3.3. The Potential Mcm1 and Sfp1 Binding Sites in the SUP35 Promoter Regions Do Not Significantly Influence Nonsense Suppression
We obtained a series of vectors based on the centromeric plasmid with the SUP35 gene pRSU1 [] with mutations and deletions in the potential Mcm1 and Sfp1 TFBSs. Using the 12-D1682 strain and the isogenic [PSI+] strains bearing the only copy of the SUP35 gene on a centromeric plasmid, we obtained both [PSI+] and [psi−] strains with SUP35 regulated by promoters with the designed mutations and deletions. Phenotypic analysis of such strains did not reveal any visible changes in the nonsense suppression phenotype (Figure 3A; Supplementary Figures S2 and S3). The only exception was the deletion of the Sfp1 TFBS in the [psi−] strain, which resulted in a slight change in color on the 1/4YEPD, indicating a slight increase in ade1-14 suppression. On several occasions we were able to observe very slow growth of this strain on media not containing adenine (Supplementary Figure S2B). We tested various [PSI+] strains for possible strain-specific allosuppression, but found none (Supplementary Figure S3). In contrast to changes in the potential Sfp1 and Mcm1 TFBSs, introducing an additional point mutation in the Abf1 TFBS close to the Mcm1-3 TFBS (Figure 2) resulted in suppression of the ade1-14 mutation (Figure 3A and Figure S2A) at levels comparable to complete deletion of the Abf1 TFBS []. Such little or no influence on nonsense suppression by the analyzed promoter mutations suggests that they do not affect SUP35 expression. To check this, we compared the Sup35 protein levels in our strains. Indeed, mutations and deletions of the potential Mcm1 TFBSs did not affect the Sup35 levels. Surprisingly, however, mutation and deletion of the Sfp1 TFBS significantly reduced Sup35 levels, though not so drastically as mutation of the Abf1 TFBS (Figure 3B and Figure S4). Thus, it is evident that potential Mcm1 TFBSs do not influence production of the Sup35 protein under normal conditions, while Sfp1 does so slightly. In case the Sfp1 TFBS is inactive, the remaining Sup35 production level is probably still sufficient for maintaining nearly normal levels of nonsense suppression.
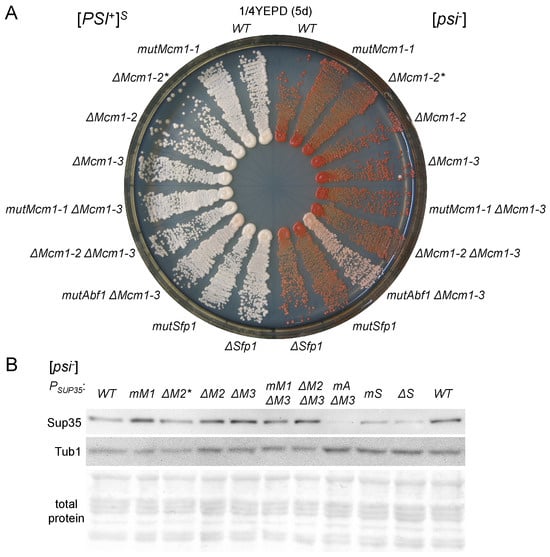
Figure 3.
The mutations in the TFBSs of Mcm1 do not affect nonsense suppression or Sup35 production. (A) Analysis of phenotypes in strains with mutations in the SUP35 promoter affecting potential Mcm1 binding sites; growth of the strains derived from U-PS-A-GT671 ([PSI+]S) and U-12-D1682 ([psi−]) on 1/4YEPD medium is shown. (B) The results of analysis of the Sup35 protein levels in U-12-D1682 derivatives performed using Western blotting. Coomassie R-250 staining was used to visualize total protein. Promoter variants are denoted as follows: WT, wild-type promoter; mM1, ΔM2, ΔM2*, ΔM3, mM1ΔM3, ΔM2ΔM3, mAΔM3, mS, and ΔS stand for mutMcm1-1, ΔMcm1-2, ΔMcm1-2*, ΔMcm1-3, mutMcm1-1ΔMcm1-3, ΔMcm1-2ΔMcm1-3, ΔMcm1-3 mutAbf1, mutSfp1, and ΔSfp1, respectively.
3.4. The Effects of Transient MCM1 Overexpression Do Not Depend on the Potential Mcm1 Binding Sites
We have shown that the potential Mcm1 TFBSs do not affect nonsense suppression when Mcm1 is produced at normal levels. However, because MCM1 overexpression enhances nonsense suppression, it is possible that additional Mcm1 may use binding sites that are not used otherwise. Thus, we tested whether the potential Mcm1 TFBSs are involved in the increase in nonsense suppression levels caused by MCM1 overexpression. We used the [PSI+] strains with mutant promoter variants and compared their suppressor phenotypes with and without overexpression of MCM1. Transient overproduction of Mcm1 resulted in slightly increased suppression in all strains, including those with double mutations in the potential TFBSs (Figure 4). Similar results were obtained in strains with other [PSI+] prion variants (Supplementary Figure S5). As the increase in suppression upon Mcm1 overproduction did not depend on the presence of functional Mcm1 TFBSs in the promoter, our results suggest that direct binding of Mcm1 to the SUP35 promoter is not involved in the effects of Mcm1 on nonsense suppression.
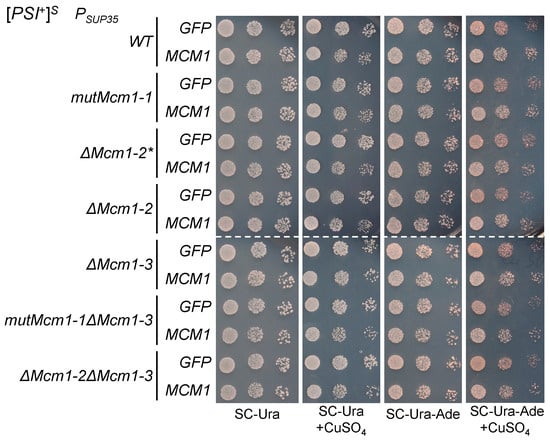
Figure 4.
Deletions and mutations of potential Mcm1 binding sites in the SUP35 promoter do not affect the suppressor effect of Mcm1 overproduction. Derivatives of the U-PS-A-GT671 strain with SUP35 under control of the mutant promoters were transformed with pRS316CG (GFP) or pU-MCM1 (MCM1). Shown are tenfold serial dilutions of the representative clones. Media were supplemented with CuSO4 at a final concentration of 50 M where indicated.
3.5. Mcm1 Does Not Form Detergent-Resistant Aggregates and Does Not Affect Sup35 Aggregation
Mcm1 is one of the Q/N-rich proteins that might be prone to aggregation, at least according to certain prediction models [,]. Taking into account that aggregates of different proteins are known to interact with each other [], a possible explanation for the effects of Mcm1 on the [PSI+] phenotype might be Mcm1 aggregation interfering with Sup35 aggregation. Thus, we first investigated whether Mcm1 could form aggregates in vivo. Transiently overproduced GFP-tagged Mcm1 demonstrated an uneven distribution, forming a single heterogeneous cluster per cell. These clusters were seen to consist of multiple small particles when viewed under high magnification (Figure 5A). This distribution pattern of Mcm1 did not depend on the presence or absence of the [PSI+] and [PIN+] prions (Supplementary Figure S6A). The clusters were presumably localized in the nucleus, which was confirmed by their colocalization when the nucleus was imaged using NLS-mCherry protein (Figure 5B). To determine whether the fluorescent foci of Mcm1-GFP corresponded to aggregates, we analyzed protein samples using semi-denaturing detergent agarose gel electrophoresis (SDD-AGE). While we were able to detect the Mcm1-GFP protein using Western blotting, its weight distribution corresponded to the monomeric protein fraction (Figure 5C), indicating that Mcm1-GFP did not form amyloid-like or any other SDS-resistant aggregates. Notably, Mcm1-GFP was stable and produced on sufficient levels (Supplementary Figure S6B).
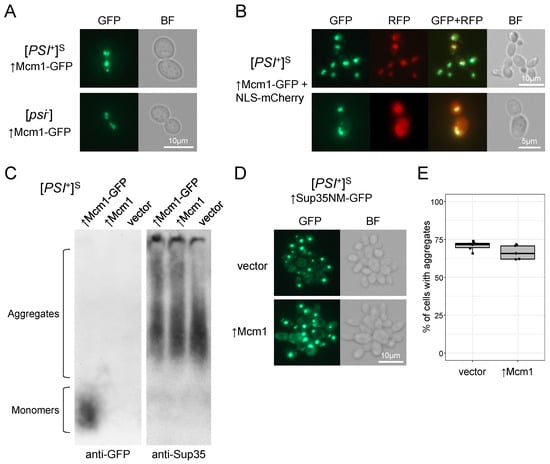
Figure 5.
Mcm1 does not form aggregates and does not influence [PSI+] aggregates. (A) Fluorescence microscopy of OT56 ([PSI+]S) and 74-D694 ([psi−]) cells transformed with the pUGC-MCM1-GFP plasmid. BF, bright field. (B) OT56 was co-transformed with pUGC-MCM1-GFP and pYX242-Nab2NLS-2mCherry and cells were analyzed with fluorescence microscopy. (C) Protein lysates of OT56 cells overproducing either Mcm1 or Mcm1-GFP were subjected to SDD-AGE and Western blotting using anti-GFP and anti-Sup35 antibodies. (D) OT56 bearing the pRS315CNMG plasmid was co-transformed with either pU-MCM1 (↑Mcm1) or pRS316 (vector). Aggregates formed by Sup35NM-GFP were visualized with fluorescence microscopy. (E) The proportions of cells with visible aggregates were counted for six vector-containing clones and five Mcm1-overproducing clones from the same transformations as in (D). No less than 50 cells were counted for each clone. Wilcoxon Mann–Whitney tests showed no significant difference between the proportions of cells with aggregates (p > 0.05). In all experiments (A–E), cells were analyzed after 4 h of copper induction.
Even though Mcm1 did not form aggregates in vivo, its overproduction could still interfere with Sup35 aggregation in [PSI+] strains. To test this, we assessed Sup35NM aggregation by co-expressing MCM1 with SUP35NM-GFP and then estimating the rate of Sup35NM aggregate appearance. We found no differences between strains overproducing Mcm1 and control strains (Figure 5D,E), indicating that excess Mcm1 does not influence [PSI+] aggregates or Sup35 aggregation. The SDD-AGE analysis also showed that Mcm1 overproduction did not alter Sup35 aggregate size distribution in [PSI+] strain (Figure 5C). In addition, no effect on the Rnq1 aggregate size distribution in the [psi−][PIN+] strain was observed (Supplementary Figure S6C). As such, it is evident that Mcm1 does not affect the [PSI+] phenotype via changes in Sup35 aggregation.
3.6. The Potential Sfp1 Binding Site in the SUP45 Promoter Affects Its Expression and Nonsense Suppression
As Sfp1 was shown to affect the transcription of both SUP35 and SUP45, we studied its potential binding sites in the promoters of both genes. First, we considered a potential TFBS in the SUP45 promoter. We introduced a mutation predicted to prevent Sfp1 binding into the centromeric plasmid pRS315-SUP45 [] and obtained strains bearing this plasmid as a sole source of the SUP45 expression. Analysis of phenotypes of the obtained strains showed a weak increase in nonsense suppression levels, as evidenced by a slight shift from red to pink color on the 1/4YEPD medium, indicating weak suppression of the ade1-14 mutation, as well as by slow growth on media lacking tryptophan, indicating suppression of trp1-289 (Figure 6A). Increased suppression implies a decrease in SUP45 expression. We checked this first by estimating the SUP45 mRNA levels using qPCR and second by assessing the Sup45 protein levels with Western blotting. Indeed, we observed a decrease in both mRNA and protein levels of SUP45 regulated by the promoter with mutant Sfp1 TFBS (Figure 6B,C). We conclude that Sfp1 acts as a transcriptional activator of SUP45 expression directly via the predicted TFBS. However, SUP45 upregulation by Sfp1 is not essential, as only slight enhancement in nonsense suppression levels is observed in its absence. The mutation in the Sfp1 TFBS did not significantly alter cell viability under normal conditions. However, excess Sfp1 is known to exacerbate lethality in a [PSI+] prion-dependent manner []. We tested for possible effects of the mutant Sfp1 TFBS on the growth inhibition caused by SFP1 overexpression, but found no influence (Supplementary Figure S7). This result corroborates the previously shown independence of the Sfp1-derived toxicity of the Sup45 abundance [].
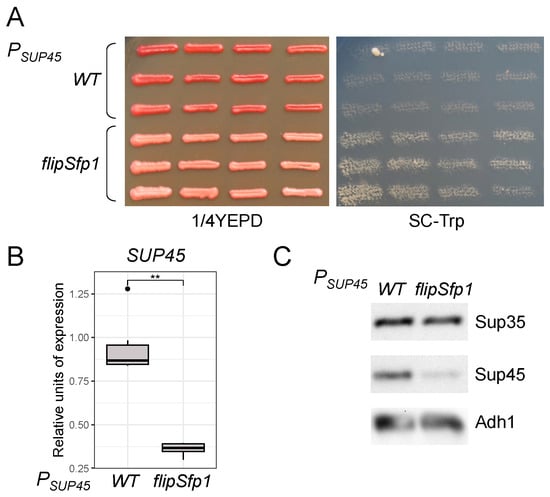
Figure 6.
Mutation of a potential Sfp1 binding site in the SUP45 promoter enhances nonsense suppression by reducing SUP45 expression. (A) Shown are twelve independently-obtained clones obtained from the U-1A-D1628 strain that bear the sole copy of the SUP45 gene under control of either wild-type (WT) or flipSfp1 promoter variant passaged on 1/4YEPD and replica plated on SC-Trp medium. (B) The results of qPCR analysis of the SUP45 mRNA levels in the strains from panel A. The relative expression of SUP45 regulated by the wild-type promoter is compared to the flipSfp1 variant. **, p < 0.01 in the Wilcoxon Mann–Whitney test. (C) Analysis of the Sup35 and Sup45 protein levels in the strains from panel A using SDS-PAGE and Western blotting. Adh1 levels were used as a reference.
3.7. The Potential Sfp1 Binding Site in the SUP35 Promoter Is Important for [PSI+] Prion Toxicity
Similar to the Sfp1 TFBS in the SUP45 promoter, we checked whether the potential Sfp1 TFBS in the SUP35 promoter had any influence on the [PSI+] prion toxicity caused by excess Sfp1. Overexpression of SFP1 appeared to be more toxic in the [PSI+] strain with the wild-type SUP35 promoter compared to strains with mutation or deletion in the Sfp1 TFBS (Figure 7A). Importantly, no such difference could be observed on the [psi−] background (Figure 7A), suggesting that the presence of the Sfp1 TFBS may contribute to the prion-dependency of the observed lethality. As we previously reported that SUP35 upregulation is one of the factors responsible for Sfp1-derived [PSI+] toxicity, we checked whether the defects in the Sfp1 TFBS affect Sup35 production. Not only did both the mutation and deletion of Sfp1 TFBS reduce the amount of the Sup35 protein, they also prevented an increase in its production during SFP1 overexpression (Figure 7B), which implies that the effect of Sfp1 on the Sup35 levels depends on the intact Sfp1 binding site in the SUP35 promoter. Consequently, the Sfp1 TFBS is responsible for the elevated SUP35 expression, which in turn enhances the toxicity in a [PSI+] prion-dependent manner. However, it should be noted that the decrease in growth during SFP1 overexpression is more pronounced in [PSI+] compared to [psi−] strains even in the absence of the Sfp1 binding site, indicating that SUP35 upregulation is not the only mechanism of the prion toxicity in this case.
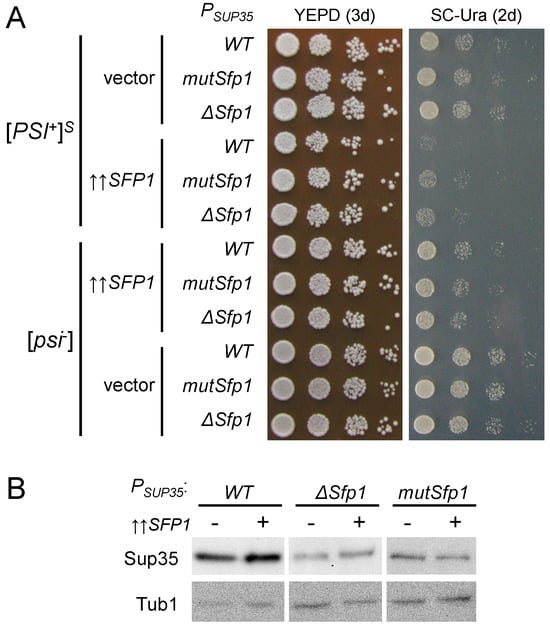
Figure 7.
SUP35 upregulation caused by excess Sfp1 contributes to [PSI+]-dependent toxicity and requires Sfp1 TFBS in the SUP35 promoter. (A) U-PS-A-GT671 ([PSI+]S) and U-12-D1682 ([psi−]) derivatives containing plasmids with the indicated SUP35 promoter variant as the sole source of SUP35 were transformed with pRS426-SFP1 (↑↑SFP1) or pRS426 (vector). Shown are tenfold serial dilutions of representative clones. (B) Assessment of the Sup35 protein levels in strains from (A) using SDS-PAGE and Western blotting. Tub1 levels were used as a reference.
4. Discussion
The influence of various Q/N-rich proteins on yeast prion propagation has been addressed in multiple studies. Primarily, this is because they are prone to either amyloid aggregation or phase separation-based inclusion formation (or both); a number of instances in which they may aid in aggregation of each other have been documented [,,]. Among the Q/N-rich proteins, transcription regulators are frequently found (reviewed in []). Several attempts ta screening for such factors affecting the [PSI+] prion or nonsense suppression have been made; however, these have yielded contradictory results. For example, SFP1 overexpression was found to enhance nonsense suppression on the [PSI+] background, but not in other test systems [,,,]. Here, in order to uncover new mechanisms of translational accuracy, we concentrated on the effects of Sfp1 and Mcm1 on the transcription of the release factor genes.
Mcm1 is a MADS-box (an acronym of Mcm1, Agamous, Deficiens, and SRF proteins) transcription factor. It operates as a dimer to directly bind DNA in a sequence-specific manner []. Mcm1 has been shown to act as both an activator and repressor of genes involved in DNA replication, cell cycle progression, mating type-specific behaviour, stress response, and other processes (reviewed in [,,,]). While able to act by itself, Mcm1 is also known to act in cooperation with various other transcription factors and thereby participate as both an activator and repressor of different regulatory modules. For example, the combinatorial complex formation with 1, 2, and Ste12 governs both up- and downregulation of mating-type specific genes [], while alternative interaction of Mcm1 with either Fkh2 or Yox1 controls cell-cycle genes expressed late in mitosis []. Mcm1 binding sites have also been shown to repeatedly appear in the promoters of ribosomal protein genes in diverse clades of Ascomycota, presumably due to an ability of Mcm1 to cooperate with another conservative transcription factor, Rap1, in activating TFIID []. Even though regulation of the release factor genes is known to be similar to the ribosomal protein genes, sequences resembling Rap1 binding site consensus are absent in the SUP35 promoter, and no direct Rap1 binding to the SUP35 promoter has been found either in vitro or in vivo []. Yet, excess Mcm1 somehow leads to an increase in nonsense suppression and enhances SUP35 transcription. Considering that Mcm1 is often seen interacting with other transcription factors, it is possible that its excess affects the activity of some other as yet unknown factors that might in turn shift the balance of proteins involved in the control of nonsense suppression efficiency, thereby indirectly affecting the [PSI+] prion phenotype.
Another possible way in which Mcm1 could influence the [PSI+] phenotype is possible interference with Sup35 aggregation. Mcm1 contains a C-terminal domain enriched in Q (42.8%) and N (6%) residues. The role of this domain in Mcm1 function is unclear, as it is not essential and its presence is not required for the functioning of Mcm1 as a transcription factor, at least within the mating type-specific regulatory network [,]. While domain requirements for other regulatory activities of Mcm1 remain unknown, considering that it uses a conservative DNA-binding motif in most of its detectable DNA-binding events [], it would be reasonable to assume that the Q/N-rich domain is also unnecessary, at least for binding promoters of other target genes. The C-terminal domain is preceded by an acidic tract composed almost exclusively of aspartate (D) and glutamate (E) residues []. Such a structure resembles that of the Sup35 protein, which contains a Q/N-rich N-terminal domain and D/E-rich part in the middle of the M-domain. Nevertheless, Sup35 forms a prion, while Mcm1 does not seem to form detergent-resistant aggregates in vivo. In a large survey of candidate prion proteins, Q/N-rich Mcm1 fragments showed no signs of aggregation either in vitro or in vivo in all tests performed [], which is consistent with our results. The reason for this may lie in the composition of the C-terminal domain of Mcm1, as in addition to Q and N it is also enriched in proline (P) residues (9.0%). The presence of prolines is thought to destabilize the amyloid structure.; even single proline substitutions in the residues within the amyloid core of Sup35 aggregates result in the loss of prion []. In the case of Mcm1, proline residues flank almost all Q-stretches; the maximum length of polyQ sequence uninterrupted by P or other residues is 10 amino acids (aa), while the longest Q/N-rich sequence between two prolines is 20 aa. Studies of polyQ aggregation in yeast suggest that such stretches are too short for amyloid formation, as polyQ sequences of 20 aa or even 25 aa have been used as no-aggregation controls in studies of aggregation of Huntingtin with larger polyQ tracts [,]. However, a 25Q-GFP Huntingtin variant was shown to form insoluble aggregates seeded by polymers of other Q/N-rich proteins, including Sup35 []. Even though a similar effect could be expected from Mcm1, we did not detect its aggregates, even in cells with [PSI+] and [PIN+] amyloids (Figure 5C). One reason might be nuclear localization of Mcm1, which makes its interaction with cytosolic prion particles highly problematic. In contrast, redirecting Huntingtin to the nucleus by addition of a nuclear localization signal (NLS) led to the formation of nuclear aggregates in the case of NLS-23Q-GFP [] but not that of NLS-20Q-GFP []. Transiently overproduced Mcm1-GFP also formed aggregate-like dots in the nucleus (Figure 5A); however, these these were not detergent-resistant, and consequently unlikely to be amyloid. It is possible that, as in the case of nuclear-localized Huntingtin, uneven nuclear distribution of Mcm1-GFP is caused by cellular protein quality control (PQC) machinery, which is known to be active inside the nucleus [,]. Thus, the puncta-like structures of Mcm1-GFP that we observed in the nucleus are likely PQC compartments rather than aggregates.
Sfp1 is an unusual Zn-finger transcription factor in which two Cys2His2-type domains are separated by a sequence of 37–39 aa. The usual distance for such proteins is 7–8 aa, leading to its name, “Split finger protein” []. Sfp1 is a nonessential transcription factor; however, its absence results in a substantial decrease in cell growth rate in combination with smaller cell size. Sfp1 is considered a key regulator of multiple processes under normal unstressed conditions. It is known to take part in the regulation of G1/S and G2/M cell cycle progression and DNA-damage response, while its main course of action is activation of the ribosomal protein (RP) and ribosome biogenesis (RiBi) genes [,,,]. Transcriptome analysis of Sfp1-deficient cells has revealed changes in expression of a large number of genes, up to 2000, which is almost third of all yeast genes [,,]. However, the slow-growth phenotype of the sfp1Δ cells and total decrease in cellular translation rates make it difficult to distinguish between direct and indirect effects. The deletion of SFP1 has been shown to reduce the expression of the SUP35 and SUP45 genes [,]; again, however, there has been no evidence that these effects are the consequences of direct regulation. Similarly, the transcription of some 2000 genes was affected in response to the overexpression of SFP1 []. The transcription of both SUP35 and SUP45 has been shown to be increased during SFP1 overexpression, even though the difference on the protein level could be detected only for Sup35 [,]. Here we present evidence of direct regulation of the SUP35 and SUP45 genes by Sfp1. Interestingly, even though the absence of functional Sfp1 TFBSs in the promoters reduces both Sup35 and Sup45 protein levels (Figure 3B and Figure 6C), excess Sfp1 affects only Sup35 []. It is possible that under normal conditions the Sup45 protein is already produced at its maximum levels, meaning that there is no capacity for additional production. This could explain why the effects of the mutations in the Sfp1 TFBSs lead to a more pronounced phenotype in the case of SUP45 (Figure 6A and Figure S2). However, this may also be due to differences in the requirements of Sup35 and Sup45 for the nonsense suppression level control. Alternatively, there might be additional feedback mechanisms for the maintenance of Sup45 but not Sup35 abundance, even though no such mechanism has been discovered yet. In the only described system in which reduction of one release factor led to decrease in another, this worked both ways; however, specific engineered promoters were used for Sup35 and Sup45 production [], so whether such feedback mechanisms exist for natively expressed genes remains unknown. Nonsense mutations in SUP35 or SUP45 do not lead to reduced levels of eRF1 or eRF3, respectively [].
The targets of Sfp1 were found to be enriched in PAC and RRPE elements [,]. The latter was later shown to correspond to the Sfp1 TFBS profile derived from ChIP-chip analysis [], which is identical to the profile in the JASPAR database that we used in this work. However, it turned out that multiple Sfp1 targets were missed by ChIP-chip and ChIP-seq. Another technique, ChEC-seq, allowed identification of Sfp1 binding to the promoters of RiBi and RiBi-like genes, suggesting several distinct modes of Sfp1 action depending on its cooperation with other transcription factors, such as Swi4 and Ifh1 []. The regulation of the expression of the release factor genes has been shown to be similar to that of the RP and RiBi genes; thus, they are also included in the RiBi-like group, even though their promoters do not contain PAC elements [,]. The promoters of RiBi-like genes have been shown to be enriched in the RRPE-like sequences. Thus, Sfp1 has been previously shown to physically interact with the SUP35 and SUP45 promoters []; however, it is not clear whether this interaction has any effect on the functioning of these genes. Here, we show that this mode of Sfp1 action makes it one of the transcription factors that balance the basic level of nonsense suppression. Another such factor is Abf1, which has also been shown to upregulate the release factor genes []. Both Abf1 and Sfp1 are non-essential for SUP35 and SUP45 expression, in the sense that the absence of upregulation driven by Abf1 or Sfp1 does not result in cell death as a sufficient amount of the release factors is still produced. However, such cells demonstrate elevated levels of nonsense suppression (Figure 3A–B and Figure 6A–C). This effect is more visible in the case of Abf1, as even point mutations in the Abf1 TFBS lead to a moderate nonsense suppression level comparable to that of complete deletion of the TFBS []. The effect on phenotype of alterations in the Sfp1 TFBSs is much less pronounced, even though a decrease in both Sup35 and Sup45 protein levels is observed.
Apart from being a transcriptional regulator, Sfp1 also contains Q/N-rich domains. Its transient overexpression was shown to lead to the appearance of detergent-resistant Sfp1 aggregates that co-localize with the Hsp40-Sis1 chaperone. Even though, Sfp1 normally resides in the nucleus, similar to Mcm1, its aggregates are localized in the cytosol. Overproduction of Sfp1 also influences aggregation of Sup35 in the [PSI+] cells, leading to an increase in the size of aggregates. Both this influence and the observed toxicity are alleviated by additional Sis1 []. As a result, Sfp1 enhances the [PSI+] prion phenotype by simultaneously upregulating SUP35 and promoting Sup35 aggregation.
5. Conclusions
In summary, we investigated the influence of two transcription factors, Mcm1 and Sfp1, both of which enhance the [PSI+] suppressor phenotype when overproduced, on [PSI+] prion properties and nonsense suppression. While the exact mechanism behind the effects of Mcm1 remains unclear, we have shown that it is unlikely to involve direct binding to the SUP35 promoter. Mcm1 did not affect Sup35 aggregation either, suggesting that it affects the [PSI+] properties indirectly. In contrast, in the case of Sfp1 we found that it is likely to directly activate transcription of both the SUP45 and SUP35 genes. The latter is an important factor that contributes to the effects of Sfp1 on [PSI+], though it is not the only mechanism behind the enhanced [PSI+] toxicity caused by excess Sfp1. Another such mechanism is an influence of Sfp1 on Sup35 aggregation [], and it seems that both mechanisms contribute equally.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15020034/s1, Figure S1: The synthetic lethality of [PSI+] with sup45 mutations is increased upon MCM1 transient overexpression. Figure S2: Alterations in the potential Mcm1 TFBSs do not affect nonsense suppression in [psi−] strains, while deletion of the potential Sfp1 binding site in the SUP35 promoter leads to an extremely small increase in the nonsense suppression. Figure S3: Mutations and deletions of the potential TFBSs of Mcm1 and Sfp1 do not affect nonsense suppression in various [PSI+] strains. Figure S4: Analysis of the Sup35 protein levels in U-PS-A-GT671 derivatives using SDS-PAGE, followed by Western blotting with anti-Sup35 antibodies. Figure S5: Deletions and mutations of potential Mcm1 binding sites in the SUP35 promoter do not affect the suppressor effect of Mcm1 overproduction. Figure S6: The [PIN+] prion does not affect Mcm1 aggregation, and Mcm1 overproduction does not affect the [PIN+] prion. Figure S7: Mutation of the potential Sfp1 binding site in the SUP45 promoter does not affect [PSI+] toxicity. Table S1: Plasmids used in this work. Table S2: Oligonucleotides used in this work. Table S3. Yeast strains used in this work. File S1: Zip archive with original images of blots.
Author Contributions
Conceptualization, A.G.M.; methodology, A.G.M. and P.B.D.; software, A.G.M. and A.S.M.; validation, A.G.M., A.S.M., P.B.D. and G.A.Z.; formal analysis, A.G.M., A.S.M. and P.B.D.; investigation, A.G.M., A.S.M. and P.B.D.; resources, A.G.M. and G.A.Z.; data curation, A.G.M.; writing—original draft preparation, A.G.M.; writing—review and editing, A.G.M., A.S.M., P.B.D. and G.A.Z.; visualization, A.G.M. and A.S.M.; supervision, G.A.Z.; project administration, G.A.Z.; funding acquisition, A.G.M. and G.A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge Saint-Petersburg State University for a research grant (project ID Pure 115624290) for supporting the part of the work related to the acquisition of a collection of strains with mutant SUP35 and SUP45 promoters. Other parts of the work were supported by the Russian Science Foundation (RSF) grant no. 23-14-00063 (https://rscf.ru/en/project/23-14-00063/ accessed on 11 March 2024).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author.
Acknowledgments
We thank Andrei A. Tsvetkov, Elena P. Efremova, Natalia A. Zaitseva, Varvara E. Ryzhkova, and Yury A. Barbitoff for help with experiments. We also thank Anton A. Nizhnikov, and Simon Alberti for plasmids. Equipment from the resource center “Development of Molecular and Cellular Technologies” of the St. Petersburg State University Scientific Park was used in the present study.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| aa | amino acids |
| bp | base pair |
| GFP | green fluorescent protein |
| ChIP | chromatin immunoprecipitation |
| ChEC | chromatin endogenous cleavage |
| 5-FOA | 5-fluoroorotic acid |
| TFBS | transcription factor binding site |
| NLS | nuclear localization signal |
| PQC | protein quality control |
| RP | ribosomal protein |
| RiBi | ribosome biogenesis |
| PCR | polymerase chain reaction |
| qPCR | quantitative polymerase chain reaction |
| SDS-PAGE | sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SDD-AGE | semi-denaturating detergent agarose gel electrophoresis |
References
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- Trubitsina, N.; Zemlyanko, O.; Moskalenko, S.; Zhouravleva, G. From past to future: Suppressor mutations in yeast genes encoding translation termination factors. Biol. Commun. 2019, 64, 89–109. [Google Scholar] [CrossRef]
- Stansfield, I.; Akhmaloka, L.E.; Tuite, M.F. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 1996, 20, 1135–1143. [Google Scholar] [CrossRef]
- Valouev, I.A.; Kushnirov, V.V.; Ter-Avanesyan, M.D. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil. Cytoskelet. 2002, 52, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Tuite, M.F. Yeast prions and their prion-forming domain. Cell 2000, 191, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Dergalev, A.A.; Alieva, M.K.; Alexandrov, A.I. Structural bases of prion variation in yeast. Int. J. Mol. Sci. 2022, 23, 5738. [Google Scholar] [CrossRef] [PubMed]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nüske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef] [PubMed]
- Zhouravleva, G.; Frolova, L.; Le Goff, X.; Le Guellec, R.; Inge-Vechtomov, S.; Kisselev, L.; Philippe, M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995, 14, 4065–4072. [Google Scholar] [CrossRef] [PubMed]
- Stansfield, I.; Jones, K.M.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Poznyakovski, A.I.; Paushkin, S.V.; Tuite, M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995, 14, 4365–4373. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Magomedova, Z.M.; Rubel, A.A.; Kondrashkina, A.M.; Inge-Vechtomov, S.G.; Galkin, A.P. [NSI+] determinant has a pleiotropic phenotypic manifestation that is modulated by SUP35, SUP45, and VTS1 genes. Curr. Genet. 2012, 58, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Ryzhova, T.A.; Volkov, K.V.; Zadorsky, S.P.; Sopova, J.V.; Inge-Vechtomov, S.G.; Galkin, A.P. Interaction of prions causes heritable traits in Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006504. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Matiiv, A.B.; Trubitsina, N.P.; Matveenko, A.G.; Barbitoff, Y.A.; Zhouravleva, G.A.; Bondarev, S.A. Amyloid and amyloid-like aggregates: Diversity and the term crisis. Biochemistry 2020, 85, 1011–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhouravleva, G.A.; Bondarev, S.A.; Trubitsina, N.P. How big is the yeast prion universe? Int. J. Mol. Sci. 2023, 24, 11651. [Google Scholar] [CrossRef] [PubMed]
- Kiktev, D.A.; Chernoff, Y.O.; Archipenko, A.V.; Zhouravleva, G.A. Identification of genes influencing synthetic lethality of genetic and epigenetic alterations in translation termination factors in yeast. Biochem. Biophys. 2011, 438, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Matveenko, A.G.; Zemlyanko, O.M.; Zhouravleva, G.A. Identification of Saccharomyces cerevisiae genes leading to synthetic lethality of prion [PSI+] with SUP45 mutations. Mol. Biol. 2013, 47, 530–537. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Belousov, M.V.; Bondarev, S.A.; Moskalenko, S.E.; Zhouravleva, G.A. Identification of new genes that affect [PSI+] prion toxicity in Saccharomyces cerevisiae yeast. Mol. Biol. 2016, 50, 710–718. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Rogoza, T.; Goginashvili, A.; Rodionova, S.; Ivanov, M.; Viktorovskaya, O.; Rubel, A.; Volkov, K.; Mironova, L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA 2010, 107, 10573–10577. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Kondrashkina, A.M.; Antonets, K.S.; Galkin, A.P. Overexpression of genes encoding asparagine-glutamine-rich transcriptional factors causes nonsense suppression in Saccharomyces cerevisiae. Russ. J. Genet. Appl. Res. 2014, 4, 122–130. [Google Scholar] [CrossRef]
- Serio, T.R.; Cashikar, A.G.; Moslehi, J.J.; Kowal, A.S.; Lindquist, S.L. Yeast prion [psi+] and its determinant, Sup35p. Methods Enzymol. 1999, 309, 649–673. [Google Scholar] [PubMed]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef] [PubMed]
- Volkov, K.V.; Aksenova, A.Y.; Soom, M.J.; Osipov, K.V.; Svitin, A.V.; Kurischko, C.; Shkundina, I.S.; Ter-Avanesyan, M.D.; Inge-Vechtomov, S.G.; Mironova, L.N. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics 2002, 160, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Moskalenko, S.E.; Chabelskaya, S.V.; Inge-Vechtomov, S.G.; Philippe, M.; Zhouravleva, G.A. Viable nonsense mutants for the essential gene SUP45 of Saccharomyces cerevisiae. BMC Mol. Biol. 2003, 4, 2. [Google Scholar] [CrossRef]
- Danilov, L.G.; Matveenko, A.G.; Ryzhkova, V.E.; Belousov, M.V.; Poleshchuk, O.I.; Likholetova, D.V.; Sokolov, P.A.; Kasyanenko, N.A.; Kajava, A.V.; Zhouravleva, G.A.; et al. Design of a new [PSI+]-no-more mutation in SUP35 with strong inhibitory effect on the [PSI+] prion propagation. Front. Mol. Neurosci. 2019, 12, 274. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef]
- Drozdova, P.B.; Tarasov, O.V.; Matveenko, A.G.; Radchenko, E.A.; Sopova, J.V.; Polev, D.E.; Inge-Vechtomov, S.G.; Dobrynin, P.V. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strains of the Peterhof. PLoS ONE 2016, 11, e0154722. [Google Scholar] [CrossRef] [PubMed]
- Matveenko, A.G.; Drozdova, P.B.; Belousov, M.V.; Moskalenko, S.E.; Bondarev, S.A.; Barbitoff, Y.A.; Nizhnikov, A.A.; Zhouravleva, G.A. SFP1-mediated prion-dependent lethality is caused by increased Sup35 aggregation and alleviated by Sis1. Genes Cells 2016, 21, 1290–1308. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Galkin, A.P.; Lewitin, E.; Chernova, T.A.; Newnam, G.P.; Belenkiy, S.M. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol. 2000, 35, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Matveenko, A.G.; Drozdova, P.B.; Moskalenko, S.E.; Tarasov, O.V.; Zhouravleva, G.A. Whole genome sequencing data and analyses of the underlying SUP35 transcriptional regulation for a Saccharomyces cerevisiae nonsense suppressor mutant. Data Brief 2019, 23, 103694. [Google Scholar] [CrossRef] [PubMed]
- Barbitoff, Y.A.; Matveenko, A.G.; Matiiv, A.B.; Maksiutenko, E.M.; Moskalenko, S.E.; Drozdova, P.B.; Polev, D.E.; Beliavskaia, A.Y.; Danilov, L.G.; Predeus, A.V.; et al. Chromosome-level genome assembly and structural variant analysis of two laboratory yeast strains from the Peterhof Genetic Collection lineage. G3 2021, 11, jkab029. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Ryzhkova, V.E.; Zaytseva, N.A.; Danilov, L.G.; Mikhailichenko, A.S.; Barbitoff, Y.A.; Zhouravleva, G.A. Processing of fluorescent proteins may prevent detection of prion particles in [PSI+] cells. Biology 2022, 11, 1688. [Google Scholar] [CrossRef] [PubMed]
- Maksiutenko, E.M.; Barbitoff, Y.A.; Matveenko, A.G.; Moskalenko, S.E.; Zhouravleva, G.A. Gene amplification as a mechanism of yeast adaptation to nonsense mutations in release factor genes. Genes 2021, 12, 2019. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kaiser, C.; Michaelis, S.; Mitchell, A. Methods in Yeast Genetics; Number 316; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1994; p. 234. [Google Scholar]
- Eaglestone, S.S.; Ruddock, L.W.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lei, J.; Yang, H.; Xu, K.; Wang, R.; Zhang, Z. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 2011, 28, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and analysis of prion and amyloid aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Lindquist, S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. JoVE 2008, 17, 838. [Google Scholar]
- Chabelskaya, S.; Kiktev, D.; Inge-Vechtomov, S.; Philippe, M.; Zhouravleva, G. Nonsense mutations in the essential gene SUP35 of Saccharomyces cerevisiae are non-lethal. Mol. Genet. Genom. MGG 2004, 272, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Kiktev, D.; Moskalenko, S.; Murina, O.; Baudin-Baillieu, A.; Rousset, J.P.; Zhouravleva, G. The paradox of viable sup45 STOP mutations: A necessary equilibrium between translational readthrough, activity and stability of the protein. Mol. Genet. Genom. MGG 2009, 282, 83–96. [Google Scholar] [CrossRef]
- Kwon, A.T.; Arenillas, D.J.; Hunt, R.W.; Wasserman, W.W. oPOSSUM-3: Advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 2012, 2, 987–1002. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Venters, B.J.; Wachi, S.; Mavrich, T.N.; Andersen, B.E.; Jena, P.; Sinnamon, A.J.; Jain, P.; Rolleri, N.S.; Jiang, C.; Hemeryck-Walsh, C.; et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 2011, 41, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Michelitsch, M.D.; Weissman, J.S. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 2000, 97, 11910–11915. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, S.A.; Antonets, K.S.; Kajava, A.V.; Nizhnikov, A.A.; Zhouravleva, G.A. Protein Co-Aggregation Related to Amyloids: Methods of Investigation, Diversity, and Classification. Int. J. Mol. Sci. 2018, 19, 2292. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Richmond, T.J. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 1998, 391, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Tye, B.K.; Chang, V.K. Dual functional regulators coordinate DNA replication and gene expression in proliferating cells. Front. Biosci. 2004, 9, 2548–2555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wittenberg, C.; Reed, S.I. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 2005, 24, 2746–2755. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, R.G.; Hooda, J.; Shah, A.; Cao, T.M.; Henke, R.M.; Zhang, L. The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci. 2012, 2, 30. [Google Scholar] [CrossRef] [PubMed]
- Dolan, J.W.; Fields, S. Cell-type-specific transcription in yeast. Biochim. Biophys. Acta Gene Struct. Expr. 1991, 1088, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Darieva, Z.; Clancy, A.; Bulmer, R.; Williams, E.; Pic-Taylor, A.; Morgan, B.A.; Sharrocks, A.D. A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol. Cell 2010, 38, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, T.R.; Johnson, A.N.; Howard, C.J.; Britton, C.S.; Fowler, K.R.; Feigerle, J.T.; Weil, P.A.; Johnson, A.D. Intrinsic cooperativity potentiates parallel cis-regulatory evolution. eLife 2018, 7, e37563. [Google Scholar] [CrossRef]
- Rossi, M.J.; Lai, W.K.M.; Pugh, B.F. Genome-wide determinants of sequence-specific DNA binding of general regulatory factors. Genome Res. 2018, 28, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Primig, M.; Winkler, H.; Ammerer, G. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 1991, 10, 4209–4218. [Google Scholar] [CrossRef]
- Bruhn, L.; Hwang-Shum, J.J.; Sprague, G.F., Jr. The N-terminal 96 residues of MCM1, a regulator of cell type-specific genes in Saccharomyces cerevisiae, are sufficient for DNA binding, transcription activation, and interaction with alpha 1. Mol. Cell. Biol. 1992, 12, 3563–3572. [Google Scholar] [PubMed]
- Chang, H.Y.; Lin, J.Y.; Lee, H.C.; Wang, H.L.; King, C.Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. USA 2008, 105, 13345–13350. [Google Scholar] [CrossRef]
- Krobitsch, S.; Lindquist, S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Muchowski, P.J.; Schaffar, G.; Sittler, A.; Wanker, E.E.; Hayer-Hartl, M.K.; Hartl, F.U. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2000, 97, 7841–7846. [Google Scholar] [CrossRef] [PubMed]
- Serpionov, G.V.; Alexandrov, A.I.; Antonenko, Y.N.; Ter-Avanesyan, M.D. A protein polymerization cascade mediates toxicity of non-pathological human huntingtin in yeast. Sci. Rep. 2015, 5, 18407. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.E.; Lo, R.S.; Davis, C.; Strand, A.D.; Neal, C.L.; Olson, J.M.; Fields, S. Altered transcription in yeast expressing expanded polyglutamine. Proc. Natl. Acad. Sci. USA 2001, 98, 13201–13206. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Levine, J.J.; Li, S.H.; Li, X.J. Nuclear aggregation of huntingtin is not prevented by deletion of chaperone Hsp104. Biochim. Biophys. Acta Mol. Basis Dis. 2001, 1537, 158–166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, S.H.; Kukushkin, Y.; Gupta, R.; Chen, T.; Konagai, A.; Hipp, M.S.; Hayer-Hartl, M.; Hartl, F.U. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 2013, 154, 134–145. [Google Scholar] [CrossRef]
- Miller, S.B.; Ho, C.T.; Winkler, J.; Khokhrina, M.; Neuner, A.; Mohamed, M.Y.; Guilbride, D.L.; Richter, K.; Lisby, M.; Schiebel, E.; et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015, 34, 778–797. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, H.; Silver, P. A split zinc-finger protein is required for normal yeast growth. Gene 1991, 107, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Norris, D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics 1998, 150, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.; Nishikawa, J.L.; Breitkreutz, B.J.; Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 2002, 297, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Fingerman, I.; Nagaraj, V.; Norris, D.; Vershon, A.K. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2003, 2, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Marion, R.M.; Regev, A.; Segal, E.; Barash, Y.; Koller, D.; Friedman, N.; O’Shea, E.K. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 2004, 101, 14315–14322. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.; Rupes, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [PubMed]
- Cipollina, C.; van den Brink, J.; Daran-Lapujade, P.; Pronk, J.T.; Porro, D.; de Winde, J.H. Saccharomyces cerevisiae SFP1: At the crossroads of central metabolism and ribosome biogenesis. Microbiology 2008, 154, 1686–1699. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Drozdova, P.; Rogoza, T.; Radchenko, E.; Lipaeva, P.; Mironova, L. Transcriptional response to the [ISP+] prion of Saccharomyces cerevisiae differs from that induced by the deletion of its structural gene, SFP1. FEMS Yeast Res. 2014, 14, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Radchenko, E.; Rogoza, T.; Khokhrina, M.; Drozdova, P.; Mironova, L. SUP35 expression is enhanced in yeast containing [ISP+], a prion form of the transcriptional regulator Sfp1. Prion 2011, 5, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, P.B.; Radchenko, E.A.; Rogoza, T.M.; Khokhrina, M.A.; Mironova, L.N. The SFP1 controls translation termination in Saccharomyces cerevisiae via regulation of Sup35p (eRF3) level. Mol. Biol. 2013, 47, 242–247. [Google Scholar] [CrossRef]
- Hughes, J.D.; Estep, P.W.; Tavazoie, S.; Church, G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000, 296, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Byers, K.J.; McCord, R.P.; Shi, Z.; Berger, M.F.; Newburger, D.E.; Saulrieta, K.; Smith, Z.; Shah, M.V.; Radhakrishnan, M.; et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009, 19, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Albert, B.; Tomassetti, S.; Gloor, Y.; Dilg, D.; Mattarocci, S.; Kubik, S.; Shore, D. Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 2019, 33, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Dagkessamanskaya, A.; Ter-Avanesyan, M.; Mager, W.H. Transcriptional regulation of SUP35 and SUP45 in Saccharomyces cerevisiae. Yeast 1997, 13, 1265–1274. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).