A Density-Based Approach for Leaf Area Index Assessment in a Complex Forest Environment Using a Terrestrial Laser Scanner
Abstract
:1. Introduction
2. Data
2.1. HAVO Site
2.2. Leaf Area Index (LAI) Measurement
2.3. The Compact Biomass Lidar (CBL)
3. Methods
3.1. Point Cloud Denoising
3.2. Detecting the Canopy
3.3. Finding Voxel Sizes
3.4. Measuring the Volume Density
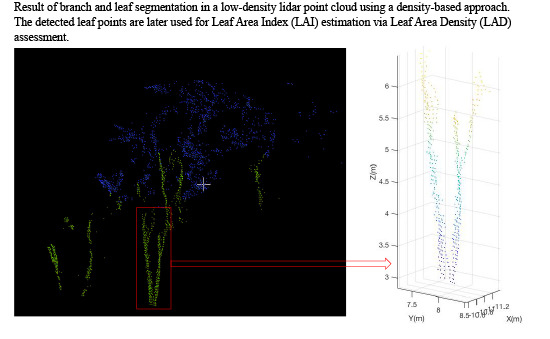
3.5. Detecting Leaf Points
KDE Bandwidth Selection
3.6. LAI Estimation Using LAD
4. Results and Discussion
4.1. Results
4.2. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, J.M.; Rich, P.M.; Gower, S.T.; Norman, J.M.; Plummer, S. Leaf area index of boreal forests: Theory, techniques, and measurements. J. Geophys. Res. Atmos. 1997, 102, 29429–29443. [Google Scholar] [CrossRef]
- Whitehead, D.; Hinckley, T.M. Models of water flux through forest stands: Critical leaf and stand parameters. Tree Physiol. 1991, 9, 35–57. [Google Scholar] [CrossRef] [PubMed]
- Amthor, J.S.; Gill, D.S.; Bormann, F.H. Autumnal leaf conductance and apparent photosynthesis by saplings and sprouts in a recently disturbed northern hardwood forest. Oecologia 1990, 84, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Davi, H.; Soudani, K.; Deckx, T.; Dufrene, E.; Le Dantec, V.; Francois, C. Estimation of forest leaf area index from SPOT imagery using NDVI distribution over forest stands. Int. J. Remote Sens. 2006, 27, 885–902. [Google Scholar] [CrossRef]
- Wilhelm, W.W.; Ruwe, K.; Schlemmer, M.R. Comparison of three leaf area index meters in a corn canopy. Crop Sci. 2000, 40, 1179–1183. [Google Scholar] [CrossRef]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- Morsdorf, F.; Kötz, B.; Meier, E.; Itten, K.I.; Allgöwer, B. Estimation of LAI and fractional cover from small footprint airborne laser scanning data based on gap fraction. Remote Sens. Environ. 2006, 104, 50–61. [Google Scholar] [CrossRef]
- Martens, S.N.; Ustin, S.L.; Rousseau, R.A. Estimation of tree canopy leaf area index by gap fraction analysis. For. Ecol. Manag. 1993, 61, 91–108. [Google Scholar] [CrossRef]
- Jarvis, P.G.; Leverenz, J.W. Productivity of temperate, deciduous and evergreen forests. In Physiological Plant Ecology IV; Springer: Berlin/Heidelberg, Germany, 1983; pp. 233–280. [Google Scholar]
- Henning, J.G.; Radtke, P.J. Detailed stem measurements of standing trees from ground-based scanning lidar. For. Sci. 2006, 52, 67–80. [Google Scholar]
- Li, S.; Dai, L.; Wang, H.; Wang, Y.; He, Z.; Lin, S. Estimating leaf area density of individual trees using the point cloud segmentation of terrestrial LiDAR data and a voxel-based model. Remote Sens. 2017, 9, 1202. [Google Scholar] [CrossRef]
- Jonckheere, I.; Fleck, S.; Nackaerts, K.; Muys, B.; Coppin, P.; Weiss, M.; Baret, F. Review of methods for in situ leaf area index determination: Part I. Theories, sensors and hemispherical photography. Agric. For. Meteorol. 2004, 121, 19–35. [Google Scholar] [CrossRef]
- Hosoi, F.; Omasa, K. Factors contributing to accuracy in the estimation of the woody canopy leaf area density profile using 3D portable lidar imaging. J. Exp. Bot. 2007, 58, 3463–3473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.L.; Fournier, R.A.; Verstraete, M.M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial LiDAR. Agric. For. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Wilson, J.W. Analysis of the spatial distribution of foliage by two-dimensional point quadrats. New Phytol. 1959, 58, 92–99. [Google Scholar] [CrossRef]
- Chen, J.M.; Black, T.A.; Adams, R.S. Evaluation of hemispherical photography for determining plant area index and geometry of a forest stand. Agric. For. Meteorol. 1991, 56, 129–143. [Google Scholar] [CrossRef]
- Omasa, K.; Hosoi, F.; Konishi, A. 3D lidar imaging for detecting and understanding plant responses and canopy structure. J. Exp. Bot. 2006, 58, 881–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, S.; Wu, F.; Guo, Q.; Wang, Y.; Li, W.; Xue, B.; Hu, X.; Li, P.; Tian, D.; Li, C.; et al. Segmenting tree crowns from terrestrial and mobile LiDAR data by exploring ecological theories. ISPRS J. Photogramm. Remote Sens. 2015, 110, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Ghebrezgabher, M.G.; Yang, T.; Yang, X.; Wang, X.; Khan, M. Extracting and analyzing forest and woodland cover change in Eritrea based on landsat data using supervised classification. Egypt. J. Remote Sens. Space Sci. 2016, 19, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.; Kang, E.; Wang, S.; Lee, K.H. A new segmentation method for point cloud data. Int. J. Mach. Tools Manuf. 2002, 42, 167–178. [Google Scholar] [CrossRef]
- Yao, W.; Hinz, S.; Stilla, U. Object extraction based on 3D-segmentation of lidar data by combining mean shift with normalized cuts: Two examples from urban areas. In Proceedings of the 2009 Joint Urban Remote Sensing Event, Shanghai, China, 20–22 May 2009. [Google Scholar]
- Kelbe, D.; van Aardt, J.; Romanczyk, P.; van Leeuwen, M.; Cawse-Nicholson, K. Single-scan stem reconstruction using low-resolution terrestrial laser scanner data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3414–3427. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Walker, L.R.; Whiteaker, L.D.; Matson, P.A. Nutrient limitations to plant growth during primary succession in Hawaii Volcanoes National Park. Biogeochemistry 1993, 23, 197–215. [Google Scholar] [CrossRef]
- Giambelluca, T.W.; Martin, R.E.; Asner, G.P.; Huang, M.; Mudd, R.G.; Nullet, M.A.; Foote, D. Evapotranspiration and energy balance of native wet montane cloud forest in Hawai‘i. Agric. For. Meteorol. 2009, 149, 230–243. [Google Scholar] [CrossRef]
- Van der Zande, D.; Hoet, W.; Jonckheere, I.; van Aardt, J.; Coppin, P. Influence of measurement set-up of ground-based LiDAR for derivation of tree structure. Agric. For. Meteorol. 2006, 141, 147–160. [Google Scholar] [CrossRef]
- SICK. LMS100/111/120/151 Laser Measurement Systems Operating Instructions; SICK AG Waldkirch: Reute, Germany, 2009. [Google Scholar]
- Van Aardt, J.A.; Kelbe, D.; Sacca, K.; Giardina, C.P.; Selmants, P.C.; Litton, C.M.; Asner, G.P. A terrestrial lidar’s assessment of climate change impacts on forest structure. In Proceedings of the Silvilaser 2017, Blacksburg, VA, USA, 10–12 October 2017. [Google Scholar]
- Rusu, R.B.; Marton, Z.C.; Blodow, N.; Dolha, M.; Beetz, M. Towards 3D point cloud based object maps for household environments. Robot. Auton. Syst. 2008, 56, 927–941. [Google Scholar] [CrossRef]
- Samet, H.; Tamminen, M. Efficient component labeling of images of arbitrary dimension represented by linear bintrees. IEEE Trans. Pattern Anal. Mach. Intell. 1988, 10, 579–586. [Google Scholar] [CrossRef]
- Douglas, E.S.; Strahler, A.; Martel, J.; Cook, T.; Mendillo, C.; Marshall, R.; Yang, X. DWEL: A dual-wavelength echidna lidar for ground-based forest scanning. In Proceedings of the 2012 IEEE International Geoscience and Remote Sensing Symposium, Munich, Germany, 22–27 July 2012; pp. 4998–5001. [Google Scholar]
- Dewez, T.J.; Girardeau-Montaut, D.; Allanic, C.; Rohmer, J. Facets: A Cloudcompare Plugin to Extract Geological Planes from Unstructured 3D Point Clouds. Int. Arch. Photogramm. Remote Sen. Spat. Inf. Sci. 2016, 41, 799–804. [Google Scholar] [CrossRef]
- Canny, J. A computational approach to edge detection. In Readings in Computer Vision; Morgan Kaufmann: Burlington, MA, USA, 1987; pp. 184–203. [Google Scholar]
- Parzen, E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962, 33, 1065–1076. [Google Scholar] [CrossRef]
- Burkhart, H.E.; Avery, T.E.; Bullock, B.P. Forest Measurements, 6th ed.; Waveland Press: Long Grove, IL, USA, 2019. [Google Scholar]
- Matheron, G. Principles of geostatistics. Econ. Geol. 1963, 58, 1246–1266. [Google Scholar] [CrossRef]
- Antonarakis, A.S.; Richards, K.S.; Brasington, J.; Muller, E. Determining leaf area index and leafy tree roughness using terrestrial laser scanning. Water Resour. Res. 2010. [Google Scholar] [CrossRef]
- Danson, F.M.; Hetherington, D.; Morsdorf, F.; Koetz, B.; Allgower, B. Three-dimensional forest cannopy structure from terrestrial laser scanning. Koukal Schneider 2006, 13, 61–65. [Google Scholar]
- Moorthy, I.; Miller, J.R.; Hu, B.; Chen, J.; Li, Q. Retrieving crown leaf area index from an individual tree using ground-based lidar data. Can. J. Remote Sens. 2008, 34, 320–332. [Google Scholar]
- Li, Y.; Guo, Q.; Su, Y.; Tao, S.; Zhao, K.; Xu, G. Retrieving the gap fraction, element clumping index, and leaf area index of individual trees using single-scan data from a terrestrial laser scanner. ISPRS J. Photogramm. Remote Sens. 2017, 130, 308–316. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Mitchell, J.J.; Levia, D.F.; Clark, P.E.; Glenn, N.F. Estimation of big sagebrush leaf area index with terrestrial laser scanning. Ecol. Indic. 2016, 61, 815–821. [Google Scholar] [CrossRef]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Greaves, H.E.; Vierling, L.A.; Eitel, J.U.; Boelman, N.T.; Magney, T.S.; Prager, C.M.; Griffin, K.L. Estimating aboveground biomass and leaf area of low-stature Arctic shrubs with terrestrial LiDAR. Remote Sens. Environ. 2015, 164, 26–35. [Google Scholar] [CrossRef]
- Zheng, G.; Ma, L.; He, W.; Eitel, J.U.; Moskal, L.M.; Zhang, Z. Assessing the contribution of woody materials to forest angular gap fraction and effective leaf area index using terrestrial laser scanning data. IEEE Trans. Geosci. Remote Sens. 2015, 54, 1475–1487. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, X.; Schull, M.A.; Román-Colón, M.O.; Yao, T.; Wang, Z.; Newnham, G.J. Measuring effective leaf area index, foliage profile, and stand height in New England forest stands using a full-waveform ground-based lidar. Remote Sens. Environ. 2011, 115, 2954–2964. [Google Scholar] [CrossRef]










| Feature | Description |
|---|---|
| operating environment (Fahrenheit) | 32-mo122 |
| Probe length (cm) | 84 |
| Number of sensors | 80 |
| PAR range (molm s) | 0–2500 |
| Resolution (molm s) | 1 |
| Minimum spatial resolution (cm) | 1 |
| Feature | Description |
|---|---|
| Range Finder | Time-of-Flight and Intensity |
| Wavelength (nm) | 905 |
| Measured Range (m) | 0.5–50 |
| Range Accuracy (mm) | ±30 |
| Resolution (degree) | 0.25 |
| Scan duration (s) | 33 |
| Weight (kg) | 3.9 |
| Beam divergence (mrad) | 15.0 |
| Minimum angular step-width (mrad) | 4.36 |
| Coverage () | 270 × 360 |
| Maximum pulse frequency (kHz) | 27 |
| Minimum (m) | Maximum (m) | Mean (m) | Stdev (m) |
|---|---|---|---|
| 0.73 | 3.46 | 1.57 | 0.62 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouzbeh Kargar, A.; MacKenzie, R.; Asner, G.P.; van Aardt, J. A Density-Based Approach for Leaf Area Index Assessment in a Complex Forest Environment Using a Terrestrial Laser Scanner. Remote Sens. 2019, 11, 1791. https://doi.org/10.3390/rs11151791
Rouzbeh Kargar A, MacKenzie R, Asner GP, van Aardt J. A Density-Based Approach for Leaf Area Index Assessment in a Complex Forest Environment Using a Terrestrial Laser Scanner. Remote Sensing. 2019; 11(15):1791. https://doi.org/10.3390/rs11151791
Chicago/Turabian StyleRouzbeh Kargar, Ali, Richard MacKenzie, Gregory P. Asner, and Jan van Aardt. 2019. "A Density-Based Approach for Leaf Area Index Assessment in a Complex Forest Environment Using a Terrestrial Laser Scanner" Remote Sensing 11, no. 15: 1791. https://doi.org/10.3390/rs11151791