Suitability of Airborne and Terrestrial Laser Scanning for Mapping Tree Crop Structural Metrics for Improved Orchard Management
Abstract
:1. Introduction
2. Study Areas and Datasets
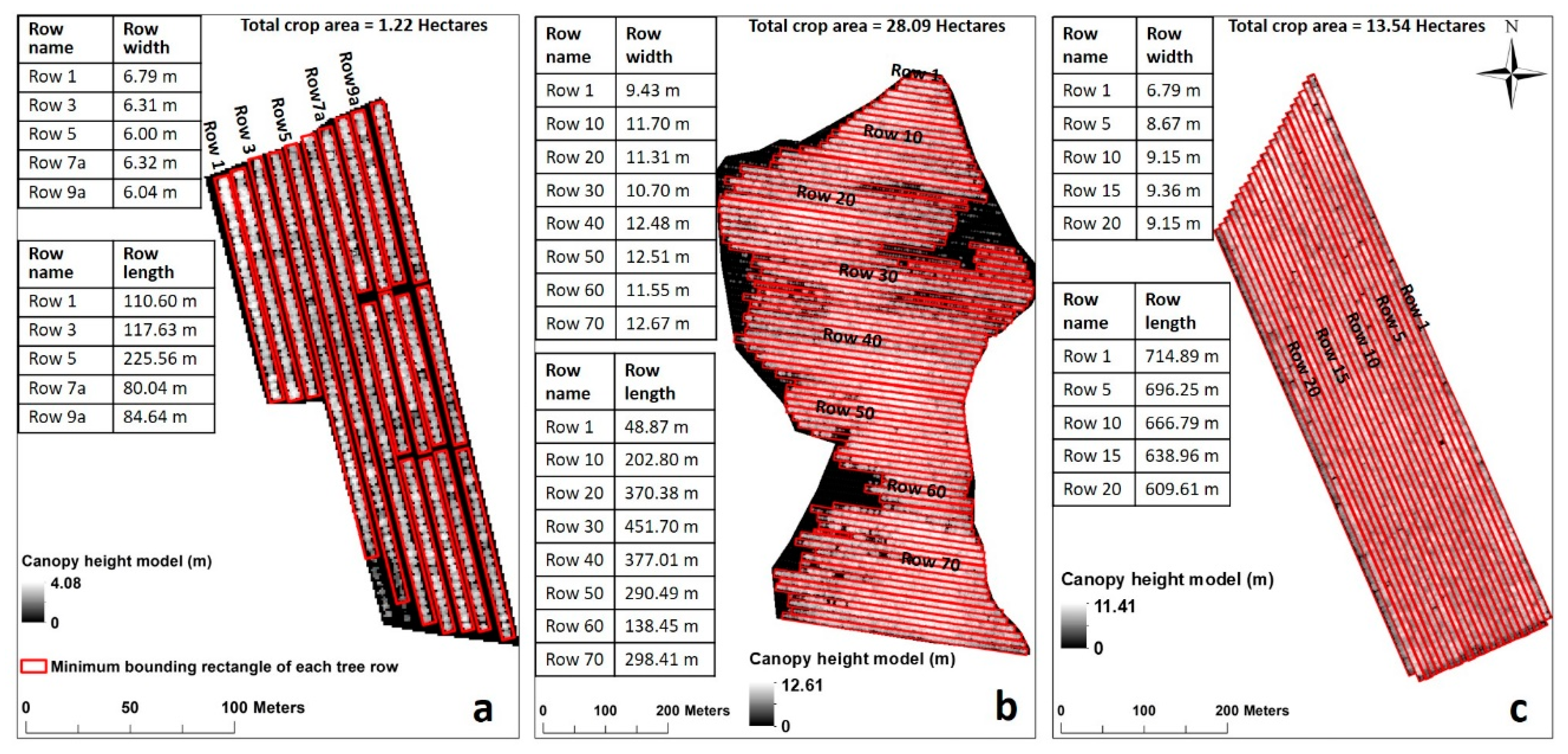
2.1. Study Area
2.2. Datasets and Methods
2.2.1. Datasets
2.2.2. Terrestrial Laser Scanning Data Processing
2.2.3. Airborne Laser Scanning Data Processing
2.2.4. Evaluation of ALS-Derived Tree Crop Structure
3. Results
3.1. Evaluation of ALS Crown Area Against TLS Data
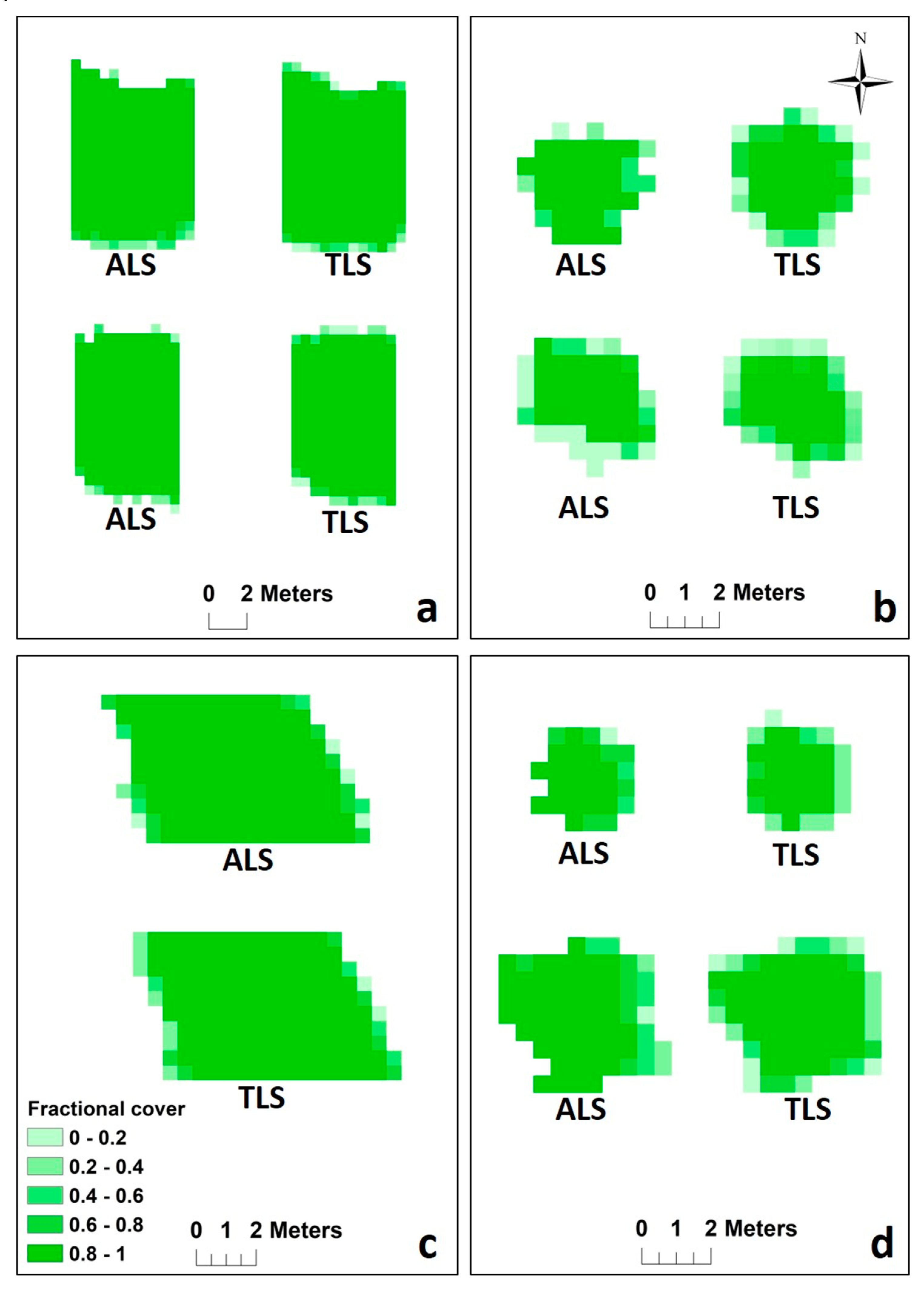
3.2. Evaluation of ALS Fractional Cover Against TLS Data
3.3. Evaluation of ALS Maximum Crown Height Against TLS Data
3.4. Evaluation of ALS Crown Volume Against TLS Data
3.5. Additional ALS Orchard and TLS Canopy Parameters
4. Discussion
4.1. Evaluating ALS-Derived Tree Crop Structure Against TLS Data
4.2. Capacity of ALS and TLS Data for Imporved Orchard Management
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shugart, H.H.; Saatchi, S.; Hall, F.G. Importance of structure and its measurement in quantifying function of forest ecosystems. J. Geophys. Res. Space Phys. 2010, 115, 115. [Google Scholar] [CrossRef]
- Hansen, C.F. Lidar Remote Sensing of Forest Canopy Structure: An Assessment of the Accuracy of Lidar and Its Relationship to Higher Trophic Levels. Master’s Thesis, University of Vermont, Burlington, VT, USA, 2015. [Google Scholar]
- Van Der Zande, D.; Hoet, W.; Jonckheere, I.; Van Aardt, J.; Coppin, P. Influence of measurement set-up of ground-based LiDAR for derivation of tree structure. Agric. Meteorol. 2006, 141, 147–160. [Google Scholar] [CrossRef]
- Rosell-Polo, J.R.; Sanz, R. A review of methods and applications of the geometric characterization of tree crops in agricultural activities. Comput. Electron. Agric. 2012, 81, 124–141. [Google Scholar] [CrossRef] [Green Version]
- Westling, F.; Underwood, J.; Örn, S. Light interception modelling using unstructured LiDAR data in avocado orchards. Comput. Electron. Agric. 2018, 153, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Phinn, S.; Johansen, K.; Robson, A.; Muir, J.; Searle, C. Estimating Changes in Leaf Area, Leaf Area Density, and Vertical Leaf Area Profile for Mango, Avocado, and Macadamia Tree Crowns Using Terrestrial Laser Scanning. Remote Sens. 2018, 10, 1750. [Google Scholar] [CrossRef] [Green Version]
- Huett, D. Macadamia physiology review: A canopy light response study and literature review. Aust. J. Agric. Res. 2004, 55, 609. [Google Scholar] [CrossRef] [Green Version]
- Mu, Y.; Fujii, Y.; Takata, D.; Zheng, B.; Noshita, K.; Honda, K.; Ninomiya, S.; Guo, W. Characterization of peach tree crown by using high-resolution images from an unmanned aerial vehicle. Hortic. Res. 2018, 5, 74. [Google Scholar] [CrossRef] [Green Version]
- Estornell, J.; Martí, B.V.; Cortés, I.L.; Salazar, D.; Fernández-Sarría, A. Estimation of wood volume and height of olive tree plantations using airborne discrete-return LiDAR data. Gisci. Remote Sens. 2014, 51, 17–29. [Google Scholar] [CrossRef]
- Hadaś, E. Accuracy of tree geometric parameters depending on the LiDAR data density. Eur. J. Remote Sens. 2016, 49, 73–92. [Google Scholar] [CrossRef] [Green Version]
- Hadas, E.; Borkowski, A.; Estornell, J.; Tymkow, P. Automatic estimation of olive tree dendrometric parameters based on airborne laser scanning data using alpha-shape and principal component analysis. Gisci. Remote Sens. 2017, 54, 898–917. [Google Scholar] [CrossRef]
- Miranda-Fuentes, A.; Llorens, J.; Gamarra-Diezma, J.L.; Gil-Ribes, J.A.; Gil, E. Towards an Optimized Method of Olive Tree Crown Volume Measurement. Sensors 2015, 15, 3671–3687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, N.; Hanan, J. Use of Functional-Structural Plant. Modelling in Horticulture; Agri-Science Queensland, Department of Agriculture, Fisheries and Forestry: Brisbane, Australia, 2012. [Google Scholar]
- Walklate, P.J.; Richardson, G.M.; Cross, J.V.; Murray, R.A. Relationship between orchard tree crop structure and performance characteristics of an axial fan sprayer. Asp. Appl. Biol. 2000, 57, 285–292. [Google Scholar]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J.; Bauhus, J. Forest and woodland stand structural complexity: Its definition and measurement. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.-L.; Fournier, R.A.; Verstraete, M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial LiDAR. Agric. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Llorens, J.; Gil, E.; Llop, J.; Queraltó, M. Georeferenced LiDAR 3D Vine Plantation Map Generation. Sensors 2011, 11, 6237–6256. [Google Scholar] [CrossRef]
- Estornell, J.; Ruiz, L.A.; Martí, B.V.; Cortés, I.L.; Salazar, D.; Fernández-Sarría, A. Estimation of pruning biomass of olive trees using airborne discrete-return LiDAR data. Biomass Bioenergy 2015, 81, 315–321. [Google Scholar] [CrossRef]
- Jang, J.; Payan, V.; Viau, A.A.; Devost, A. The use of airborne lidar for orchard tree inventory. Int. J. Remote Sens. 2008, 29, 1767–1780. [Google Scholar] [CrossRef]
- Fernández-Sarría, A.; López-Cortés, I.; Estornell, J.; Martí, B.V.; Salazar, D. Estimating residual biomass of olive tree crops using terrestrial laser scanning. Int. J. Appl. Earth Obs. Geoinf. 2019, 75, 163–170. [Google Scholar] [CrossRef]
- Moorthy, I.; Miller, J.R.; Jimenez-Berni, J.A.; Zarco-Tejada, P.J.; Hu, B.; Chen, J. Field characterization of olive (Olea europaea L.) tree crown architecture using terrestrial laser scanning data. Agric. Meteorol. 2011, 151, 204–214. [Google Scholar] [CrossRef]
- Chen, J.M.; Rich, P.M.; Gower, S.T.; Norman, J.M.; Plummer, S. Leaf area index of boreal forests: Theory, techniques, and measurements. J. Geophys. Res. Space Phys. 1997, 102, 29429–29443. [Google Scholar] [CrossRef]
- Estornell, J.; Velázquez-Martí, A.; Fernández-Sarría, A.; Cortés, I.L.; Martí-Gavilá, J.; Salazar, D. Estimación de parámetros de estructura de nogales utilizando láser escáner terrestre. Rev. Teledetección 2017, 48, 67. [Google Scholar] [CrossRef]
- Lovell, J.; Jupp, D.; Culvenor, D.S.; Coops, N.C. Using airborne and ground-based ranging lidar to measure canopy structure in Australian forests. Can. J. Remote Sens. 2003, 29, 607–622. [Google Scholar] [CrossRef]
- Hilker, T.; Van Leeuwen, M.; Coops, N.C.A.; Wulder, M.; Newnham, G.; Jupp, D.; Culvenor, D.S. Comparing canopy metrics derived from terrestrial and airborne laser scanning in a Douglas-fir dominated forest stand. Trees 2010, 24, 819–832. [Google Scholar] [CrossRef]
- Korhonen, L.; Vauhkonen, J.; Virolainen, A.; Hovi, A.; Korpela, I. Estimation of tree crown volume from airborne lidar data using computational geometry. Int. J. Remote Sens. 2013, 34, 7236–7248. [Google Scholar] [CrossRef]
- Bundaberg Fruit & Vegetable Growers. Available online: https://www.bfvg.com.au/ (accessed on 15 October 2019).
- Bureau of Meteorology. Climate Statistics for Australian Locations—Monthly Climate Statistics. Available online: http://www.bom.gov.au/climate/averages/tables/cw_039128.shtml (accessed on 11 February 2020).
- Queensland Government. Mangoes. Available online: https://www.daf.qld.gov.au/business-priorities/agriculture/plants/fruit-vegetable/fruit-vegetable-crops/mangoes (accessed on 19 May 2020).
- Horticulture Innovation Australia. Find Information, Publications, Industry Contacts and More on the Avocado Industry. Available online: http://horticulture.com.au/grower-focus/avocado/ (accessed on 11 July 2016).
- Avocados Australia. Australian Avos in Your Burger and on Your Pizza. Available online: http://industry.avocado.org.au/NewsItem.aspx?NewsId=51 (accessed on 11 July 2016).
- Australia Macadamia Society. The Australian Macadamia Industry; Australia Macadamia Society: Lismore, Australia, 2016. [Google Scholar]
- Beland, M.; Widlowski, J.-L.; Fournier, R.A. A model for deriving voxel-level tree leaf area density estimates from ground-based LiDAR. Environ. Model. Softw. 2014, 51, 184–189. [Google Scholar] [CrossRef]
- Beland, M.; Widlowski, J.-L.; Fournier, R.A.; Côté, J.-F.; Verstraete, M. Estimating leaf area distribution in savanna trees from terrestrial LiDAR measurements. Agric. Meteorol. 2011, 151, 1252–1266. [Google Scholar] [CrossRef]
- Wu, D.; Johansen, K.; Phinn, S.; Robson, A.; Tu, Y.-H. Inter-comparison of remote sensing platforms for height estimation of mango and avocado tree crowns. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102091. [Google Scholar] [CrossRef]
- Esri. What is a TIN surface? Available online: http://desktop.arcgis.com/en/arcmap/latest/manage-data/tin/fundamentals-of-tin-surfaces.htm (accessed on 19 May 2020).
- Armston, J.; Danaher, T.J.; Scarth, P.; Moffiet, T.N.; Denham, R.J. Prediction and validation of foliage projective cover from Landsat-5 TM and Landsat-7 ETM+ imagery. J. Appl. Remote Sens. 2009, 3, 033540. [Google Scholar] [CrossRef]
- Liu, L.; Pang, Y.; Li, Z.; Si, L.; Liao, S. Combining Airborne and Terrestrial Laser Scanning Technologies to Measure Forest Understorey Volume. Forests 2017, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Hancock, S.; Anderson, K.; Disney, M.; Gaston, K.J. Measurement of fine-spatial-resolution 3D vegetation structure with airborne waveform lidar: Calibration and validation with voxelised terrestrial lidar. Remote Sens. Environ. 2017, 188, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Monserud, R.A.; Huang, S. An evaluation of diagnostic tests and their roles in validating forest biometric models. Can. J. Res. 2004, 34, 619–629. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Hyyppä, H.; Maltamo, M. Effects of flight altitude on tree height estimation using airborne laser scanning. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 8, 96–101. [Google Scholar]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Pan, Y.; Hyyppä, J. Is field-measured tree height as reliable as believed—A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. Isprs J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Krooks, A.; Kaasalainen, S.; Kankare, V.; Joensuu, M.; Raumonen, P.; Kaasalainen, M. Tree structure vs. height from terrestrial laser scanning and quantitative structure models. Silva. Fenn. 2014, 48, 2. [Google Scholar] [CrossRef] [Green Version]
- Lordan, J.; Pascual, M.; Fonseca, F.; Montilla, V.; Papió, J.; Rufat, J.; Villar, J.M. An Image-based Method to Study the Fruit Tree Canopy and the Pruning Biomass Production in a Peach Orchard. HortScience 2015, 50, 1809–1817. [Google Scholar] [CrossRef]
- Jiménez-Brenes, F.M.; López-Granados, F.; De Castro, A.I.; Torres-Sánchez, J.; Serrano, N.; Peña-Barragan, J.M. Quantifying pruning impacts on olive tree architecture and annual canopy growth by using UAV-based 3D modelling. Plant. Methods 2017, 13, 55. [Google Scholar] [CrossRef] [Green Version]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; De Tanago, J.G.; Bartholomeus, H.; Brede, B.; Herold, M. Data acquisition considerations for Terrestrial Laser Scanning of forest plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- Puente, I.; González-Jorge, H.; Martínez-Sánchez, J.; Arias, P. Review of mobile mapping and surveying technologies. Measurement 2013, 46, 2127–2145. [Google Scholar] [CrossRef]
- Underwood, J.; Hung, C.; Whelan, B.; Sukkarieh, S. Mapping almond orchard canopy volume, flowers, fruit and yield using lidar and vision sensors. Comput. Electron. Agric. 2016, 130, 83–96. [Google Scholar] [CrossRef]
- Kragh, M.; Underwood, J. Multimodal obstacle detection in unstructured environments with conditional random fields. J. Field Robot. 2019, 37, 53–72. [Google Scholar] [CrossRef] [Green Version]
- Johansen, K.; Raharjo, T.; McCabe, M.F. Using Multi-Spectral UAV Imagery to Extract Tree Crop Structural Properties and Assess Pruning Effects. Remote Sens. 2018, 10, 854. [Google Scholar] [CrossRef] [Green Version]
- Sola-Guirado, R.R.; Castillo-Ruiz, F.J.; Jiménez-Jiménez, F.; Blanco-Roldan, G.L.; Castro-Garcia, S.; Gil-Ribes, J.A. Olive Actual “on Year” Yield Forecast Tool Based on the Tree Canopy Geometry Using UAS Imagery. Sensors 2017, 17, 1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayram, B.; Şeker, D.Z.; Jamil, A.; Reis, H.C.; Demir, N.; Bozkurt, S.; Ince, A.; Kucuk, T. Automatic extraction of sparse trees from high-resolution ortho-images. Arab. J. Geosci. 2018, 11, 319. [Google Scholar] [CrossRef]
- Rahman, M.M.; Robson, A.; Bristow, M. Exploring the Potential of High Resolution WorldView-3 Imagery for Estimating Yield of Mango. Remote Sens. 2018, 10, 1866. [Google Scholar] [CrossRef] [Green Version]
- Johansen, K.; Duan, Q.; Tu, Y.; Searle, C.; Wu, D.; Phinn, S.; Robson, A.; McCabe, M.F. Mapping the condition of macadamia tree crops using multi-spectral uav and worldview-3 imagery. Isprs J. Photogramm. Remote Sens. 2020, 165, 28–40. [Google Scholar] [CrossRef]
- Tickle, P.; Lee, A.; Lucas, R.; Austin, J.; Witte, C. Quantifying Australian forest floristics and structure using small footprint LiDAR and large scale aerial photography. Ecol. Manag. 2006, 223, 379–394. [Google Scholar] [CrossRef]
- Morsdorf, F.; Kötz, B.; Meier, E.; Itten, K.; Allgöwer, B. Estimation of LAI and fractional cover from small footprint airborne laser scanning data based on gap fraction. Remote Sens. Environ. 2006, 104, 50–61. [Google Scholar] [CrossRef]
- Korhonen, L.; Korpela, I.; Heiskanen, J.; Maltamo, M. Airborne discrete-return LIDAR data in the estimation of vertical canopy cover, angular canopy closure and leaf area index. Remote Sens. Environ. 2011, 115, 1065–1080. [Google Scholar] [CrossRef]
- Johansen, K.; Arroyo, L.; Armston, J.; Phinn, S.; Witte, C. Mapping riparian condition indicators in a sub-tropical savanna environment from discrete return LiDAR data using object-based image analysis. Ecol. Indic. 2010, 10, 796–807. [Google Scholar] [CrossRef]
- Trout, T.; Johnson, L.F.; Gartung, J. Remote Sensing of Canopy Cover in Horticultural Crops. HortScience 2008, 43, 333–337. [Google Scholar] [CrossRef] [Green Version]
- Majd, A.M.S.; Bleiweiss, M.P.; Dubois, D.; Shukla, M.K. Estimation of the fractional canopy cover of pecan orchards using Landsat 5 satellite data, aerial imagery, and orchard floor photographs. Int. J. Remote Sens. 2013, 34, 5937–5952. [Google Scholar] [CrossRef]
- O’connell, M.; Goodwin, I. Spatial variation of tree cover in peach orchards. Acta Hortic. 2005, 694, 203–205. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, N.; Ayars, J.E.; Phene, C.J. Biophysical response of young pomegranate trees to surface and sub-surface drip irrigation and deficit irrigation. Irrig. Sci. 2017, 35, 425–435. [Google Scholar] [CrossRef]
- Wulder, M.A.; Ortlepp, S.M.; White, J.C.; Coops, N.C.; Coggins, S.B. Monitoring tree-level insect population dynamics with multi-scale and multi-source remote sensing. J. Spat. Sci. 2008, 53, 49–61. [Google Scholar] [CrossRef]
- Menzel, C.M.; Le Lagadec, M.D. Increasing the productivity of avocado orchards using high-density plantings: A review. Sci. Hortic. 2014, 177, 21–36. [Google Scholar] [CrossRef]
- Davenport, T.L. Pruning Strategies to Maximize Tropical Mango Production From the Time of Planting to Restoration of Old Orchards. HortScience 2006, 41, 544–548. [Google Scholar] [CrossRef] [Green Version]
- Meurant, N.; Kernot, I. Mango Information Kit; Department of Primary Industries: Orange, Australia, 1999. [Google Scholar]
- Goodwin, N.R.; Coops, N.C.; Culvenor, D.S. Assessment of forest structure with airborne LiDAR and the effects of platform altitude. Remote Sens. Environ. 2006, 103, 140–152. [Google Scholar] [CrossRef]
- Tu, Y.-H.; Johansen, K.; Phinn, S.; Robson, A. Measuring Canopy Structure and Condition Using Multi-Spectral UAS Imagery in a Horticultural Environment. Remote Sens. 2019, 11, 269. [Google Scholar] [CrossRef] [Green Version]
- Tu, Y.-H.; Phinn, S.; Johansen, K.; Robson, A.; Johansen, K. Optimising drone flight planning for measuring horticultural tree crop structure. Isprs J. Photogramm. Remote Sens. 2020, 160, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Kükenbrink, D.; Schneider, F.D.; Leiterer, R.; Schaepman, M.E.; Morsdorf, F. Quantification of hidden canopy volume of airborne laser scanning data using a voxel traversal algorithm. Remote Sens. Environ. 2017, 194, 424–436. [Google Scholar] [CrossRef]
- Escola, A.; Martínez-Casasnovas, J.A.; Rufat, J.; Arnó, J.; Arbonés, A.; Sebé, F.; Pascual, M.; Gregorio, E.; Rosell-Polo, J.R. Mobile terrestrial laser scanner applications in precision fruticulture/horticulture and tools to extract information from canopy point clouds. Precis. Agric. 2016, 18, 111–132. [Google Scholar] [CrossRef] [Green Version]
- Colaço, A.; Trevisan, R.; Molin, J.P.; Rosell-Polo, J.R.; Escolà, A. Orange tree canopy volume estimation by manual and LiDAR-based methods. Adv. Anim. Biosci. 2017, 8, 477–480. [Google Scholar] [CrossRef] [Green Version]
- Henning, J.G.; Radtke, P.J. Detailed stem measurements of standing trees from ground-based scanning Lidar. For. Sci. 2006, 52, 67–80. [Google Scholar]
- Furness, G.; Magarey, P.; Miller, P.; Drew, H. Fruit tree and vine sprayer calibration based on canopy size and length of row: Unit canopy row method. Crop. Prot. 1998, 17, 639–644. [Google Scholar] [CrossRef]
- Hosoi, F.; Omasa, K. Voxel-Based 3-D Modeling of Individual Trees for Estimating Leaf Area Density Using High-Resolution Portable Scanning Lidar. IEEE Trans. Geosci. Remote Sens. 2006, 44, 3610–3618. [Google Scholar] [CrossRef]
- Glenn, N.F.; Spaete, L.; Sankey, T.; Derryberry, D.; Hardegree, S.; Mitchell, J. Errors in LiDAR-derived shrub height and crown area on sloped terrain. J. Arid. Environ. 2011, 75, 377–382. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña-Barragan, J.M. High-Throughput 3-D Monitoring of Agricultural-Tree Plantations with Unmanned Aerial Vehicle (UAV) Technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef] [Green Version]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Forest understory trees can be segmented accurately within sufficiently dense airborne laser scanning point clouds. Sci. Rep. 2017, 7, 6770. [Google Scholar] [CrossRef] [Green Version]
- Hadas, E.; Jozkow, G.; Walicka, A.; Borkowski, A. Determining geometric parameters of agricultural trees from laser scanning data obtained with unmanned aerial vehicle. Isprs Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, 407–410. [Google Scholar] [CrossRef] [Green Version]
- Hadas, E.; Jozkow, G.; Walicka, A.; Borkowski, A. Apple orchard inventory with a LiDAR equipped unmanned aerial system. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101911. [Google Scholar] [CrossRef]







| RIEGL VZ-400 TLS Scanner | RIEGL LMS-Q 1560 ALS Scanner | |
|---|---|---|
| Beam divergence | 0.35 mrad | <0.25 mrad |
| Pulse repetition rate | 300 kHz | 400 kHz |
| Laser wavelength | 1550 nm | 1064 nm |
| Minimum range | 1.5 m | 50 m |
| Maximum range | 160 m (at 20% target reflectance) 350 m (at 90% target reflectance) | 3500 m (at 20% target reflectance) 5100 m (at 60% target reflectance) |
| Field of view | 0°–360° (azimuth range) 30°–130° (zenith range) | 58° |
| Recorded data | Full waveform & up to four returns per emitted pulse | Full waveform & up to seven returns per emitted pulse |
| Accuracy | ±5 mm | ±20 mm |
| n | Max Frac Cover Difference | Min Frac Cover Difference | Average ALS Frac Cover | Average ALS Frac Cover | RMSE | |
|---|---|---|---|---|---|---|
| Avocado tree 1 | 198 | 0.84 | 0 | 0.92 | 0.93 | 0.12 |
| Avocado tree 2 | 230 | 0.84 | 0 | 0.93 | 0.89 | 0.22 |
| Mango high vigour | 71 | 0.92 | 0 | 0.78 | 0.78 | 0.33 |
| Mango low vigour | 34 | 0.88 | 0 | 0.73 | 0.72 | 0.30 |
| Macadamia tree | 163 | 1 | 0 | 0.88 | 0.87 | 0.32 |
| Small avocado tree 1 | 40 | 1 | 0 | 0.69 | 0.71 | 0.34 |
| Small avocado tree 2 | 39 | 1 | 0 | 0.66 | 0.67 | 0.30 |
| n | Max Height Difference (m) | Min Height Difference (m) | Average ALS Height (m) | Average TLS Height (m) | RMSE (m) | |
|---|---|---|---|---|---|---|
| Avocado tree 1 | 198 | 6.21 | 0.004 | 6.88 | 7.33 | 1.12 |
| Avocado tree 2 | 230 | 7.07 | 0.001 | 7.24 | 7.63 | 0.78 |
| Mango high vigour | 71 | 3.27 | 0.012 | 2.54 | 3.20 | 0.80 |
| Mango low vigour | 34 | 1.31 | 0.022 | 1.87 | 1.25 | 0.84 |
| Macadamia tree | 163 | 7.28 | 0.032 | 8.02 | 8.96 | 1.54 |
| Small avocado tree 1 | 40 | 1.64 | 0.040 | 2.47 | 2.93 | 0.59 |
| Small avocado tree 2 | 39 | 1.51 | 0.062 | 2.21 | 2.71 | 0.64 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.; Johansen, K.; Phinn, S.; Robson, A. Suitability of Airborne and Terrestrial Laser Scanning for Mapping Tree Crop Structural Metrics for Improved Orchard Management. Remote Sens. 2020, 12, 1647. https://doi.org/10.3390/rs12101647
Wu D, Johansen K, Phinn S, Robson A. Suitability of Airborne and Terrestrial Laser Scanning for Mapping Tree Crop Structural Metrics for Improved Orchard Management. Remote Sensing. 2020; 12(10):1647. https://doi.org/10.3390/rs12101647
Chicago/Turabian StyleWu, Dan, Kasper Johansen, Stuart Phinn, and Andrew Robson. 2020. "Suitability of Airborne and Terrestrial Laser Scanning for Mapping Tree Crop Structural Metrics for Improved Orchard Management" Remote Sensing 12, no. 10: 1647. https://doi.org/10.3390/rs12101647







