Measuring Alpha and Beta Diversity by Field and Remote-Sensing Data: A Challenge for Coastal Dunes Biodiversity Monitoring
Abstract
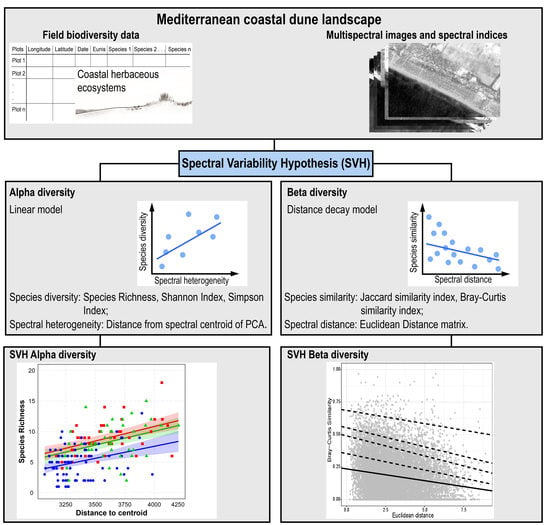
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection and Analysis
2.2.1. Data Collection
Field Data Collection
Remote-Sensing Data
2.2.2. Alpha Diversity Analysis
2.2.3. Beta Diversity Analysis
2.2.4. Spectral Variability Hypothesis Test
3. Results
3.1. Spectral Variability Hypothesis (SVH) Alpha Diversity
3.2. SVH Beta Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ceballos, G.; Ehrlich, P.R.; Barnosky, A.D.; García, A.; Pringle, R.M.; Palmer, T.M. Accelerated modern human–induced species losses: Entering the sixth mass extinction. Sci. Adv. 2015, 1, e1400253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz, S.; Demissew, S.; Carabias, J.; Joly, C.; Lonsdale, M.; Ash, N.; Larigauderie, A.; Adhikari, J.R.; Arico, S.; Báldi, A.; et al. The IPBES conceptual framework—connecting nature and people. Curr. Opin. Environ. Sustain. 2015, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Vihervaara, P.; Auvinen, A.P.; Mononen, L.; Törmä, M.; Ahlroth, P.; Anttila, S.; Böttcher, K.; Forsius, M.; Heino, J.; Heliola, J.; et al. How essential biodiversity variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 2017, 10, 43–59. [Google Scholar] [CrossRef]
- Petrou, Z.I.; Manakos, I.; Stathaki, T. Remote sensing for biodiversity monitoring: A review of methods for biodiversity indicator extraction and assessment of progress towards international targets. Biodivers. Conserv. 2015, 24, 2333–2363. [Google Scholar] [CrossRef]
- Del Vecchio, S.; Fantinato, E.; Silan, G.; Buffa, G. Trade-offs between sampling effort and data quality in habitat monitoring. Biodivers. Conserv. 2019, 28, 55–73. [Google Scholar] [CrossRef] [Green Version]
- Maccherini, S.; Bacaro, G.; Tordoni, E.; Bertacchi, A.; Castagnini, P.; Foggi, B.; Gennai, M.; Mugnai, M.; Sarmati, S.; Angiolini, C. Enough is enough? Searching for the optimal sample size to monitor european habitats: A case study from coastal sand dunes. Diversity 2020, 12, 138. [Google Scholar] [CrossRef] [Green Version]
- Rocchini, D.; Salvatori, N.; Beierkuhnlein, C.; Chiarucci, A.; de Boissieu, F.; Förster, M.; Garzon-Lopez, C.X.; Gillespie, T.W.; Hauffe, H.C.; He, K.S.; et al. From local spectral species to global spectral communities. a benchmark for ecosystem diversity estimate by remote sensing. Ecol. Inform. 2021, 61, 101195. [Google Scholar] [CrossRef]
- Rocchini, D.; Boyd, D.S.; Féret, J.B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Torresani, M.; Rocchini, D.; Sonnenschein, R.; Zebisch, M.; Marcantonio, M.; Ricotta, C.; Tonon, G. Estimating tree species diversity from space in an alpine conifer forest: The Rao’s Q diversity index meets the spectral variation hypothesis. Ecol. Inform. 2019, 52, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Torresani, M.; Rocchini, D.; Sonnenschein, R.; Zebisch, M.; Hauffe, H.C.; Heym, M.; Pretzsch, H.; Tonon, G. Height variation hypothesis: A new approach for estimating forest species diversity with CHM LiDAR data. Ecol. Indic. 2020, 117, 106520. [Google Scholar] [CrossRef]
- Rocchini, D.; Hernández-Stefanoni, J.L.; He, K.S. Advancing species diversity estimate by remotely sensed proxies: A conceptual review. Ecol. Inform. 2015, 25, 22–28. [Google Scholar] [CrossRef]
- Lausch, A.; Heurich, M.; Magdon, P.; Rocchini, D.; Schulz, K.; Bumberger, J.; King, D.J. A Range of Earth Observation Techniques for Assessing Plant Diversity. In Remote Sensing of Plant Biodiversity, 1st ed.; Cavender-Bares, J., Gamon, J., Townsend, P., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 309–348. [Google Scholar]
- Rocchini, D. Effects of spatial and spectral resolution in estimating ecosystem α-diversity by satellite imagery. Remote Sens. Environ. 2007, 111, 423–434. [Google Scholar] [CrossRef]
- Rocchini, D. Distance decay in spectral space in analyzing β-diversity. Int. J. Remote Sens. 2007, 28, 2635–2644. [Google Scholar] [CrossRef]
- Nagendra, H.; Rocchini, D.; Ghate, R.; Sharma, B.; Pareeth, S. Assessing plant diversity in a dry tropical forest: Comparing the utility of Landsat and Ikonos satellite images. Remote Sens. 2010, 2, 478–496. [Google Scholar] [CrossRef] [Green Version]
- Heumann, B.W.; Hackett, R.A.; Monfils, A.K. Testing the spectral diversity hypothesis using spectroscopy data in a simulated wetland community. Ecol. Inform. 2015, 25, 29–34. [Google Scholar] [CrossRef]
- Hall, K.; Reitalu, T.; Sykes, M.T.; Prentice, H.C. Spectral heterogeneity of QuickBird satellite data is related to fine-scale plant species spatial turnover in semi-natural grasslands. Appl. Veg. Sci. 2012, 15, 145–157. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Ramoelo, A.; Mutanga, O. Remote sensing of species diversity using Landsat 8 spectral variables. ISPRS J. Photogramm. 2017, 133, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, S.; Schmitt, T.M.; Chiarucci, A.; Irl, S.D.H.; Rocchini, D.; Vetaas, O.R.; Tanase, M.A.; Stéphane, M.; Bouvet, A.; Beierkuhnlein, C. Remote sensing of β-diversity: Evidence from plant communities in a semi-natural system. Appl. Veg. Sci. 2019, 22, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Rocchini, D.; Nagendra, H.; Ghate, R.; Cade, B.S. Spectral distance decay: Assessing species beta-diversity by quantile regression. Photogramm. Eng. Remote Sens. 2009, 75, 1225–1230. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.182.369&rep=rep1&type=pdf (accessed on 14 May 2021).
- Féret, J.B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Latifi, H.; Rossi, S. Forest beta-diversity analysis by remote sensing: How scale and sensors affect the Rao’s Q index. Ecol. Indic. 2019, 106, 105520. [Google Scholar] [CrossRef]
- Gholizadeh, H.; Gamon, J.A.; Helzet, C.J.; Cavender-Bares, J. Multi-temporal assessment of grassland α- and β-diversity using hyperspectral imaging. Ecol. Appl. 2020, 30, e02145. [Google Scholar] [CrossRef]
- Schmidtlein, S.; Fassnacht, F.E. The spectral variability hypothesis does not hold across landscapes. Remote Sens. Environ. 2017, 192, 114–125. [Google Scholar] [CrossRef] [Green Version]
- Martínez, M.L.; Psuty, N.P. Coastal Dunes. Ecology and Conservation, 1st ed.; Springer: Berlin, Germany, 2004; pp. 3–29. [Google Scholar]
- Hesp, P. A Ecological processes and plant adaptations on coastal dunes. J. Arid Environ. 1991, 21, 165–191. [Google Scholar] [CrossRef]
- Acosta, A.T.R.; Blasi, C.; Carranza, M.L.; Ricotta, C.; Stanisci, A. Quantifying ecological mosaic connectivity with a new topoecological index. Phytocoenologia 2003, 33, 623–631. [Google Scholar] [CrossRef]
- Kim, D.; Yu, K.B. A conceptual model of coastal dune ecology synthesizing spatial gradients of vegetation, soil, and geomorphology. Plant Ecol. 2009, 202, 135–148. [Google Scholar] [CrossRef]
- Bazzichetto, M.; Malavasi, M.; Acosta, A.T.R.; Carranza, M.L. How does dune morphology shape coastal EC habitats occurrence? A remote sensing approach using airborne LiDAR on the Mediterranean coast. Ecol. Indic. 2016, 71, 618–626. [Google Scholar] [CrossRef]
- McLachlan, A.; Brown, A.C. The Ecology of Sandy Shores, 2nd ed.; Elsevier: Burlington, NJ, USA, 2006; pp. 273–323. [Google Scholar]
- Doody, J.P. Sand Dune Conservation, Management and Restoration, 1st ed.; Springer Science: Dordrecht, The Netherlands, 2013; pp. 37–82. [Google Scholar]
- Janssen, J.A.M.; Rodwell, J.S.; García Criado, M.; Gubbay, S.; Haynes, T.; Nieto, A.; Sanders, N.; Landucci, F.; Loidi, J.; Ssymank, A.; et al. European Red List of Habitats. Part 2. Terrestrial and Freshwater Habitats, 1st ed.; European Commision: Luxembourg, 2016; pp. 1–40. [Google Scholar]
- Prisco, I.; Angiolini, C.; Assini, S.; Buffa, G.; Gigante, D.; Marcenó, C.; Sciandrello, S.; Villani, M.; Acosta, A.T.R. Conservation status of Italian coastal dune habitats in the light of the 4th monitoring report (92/43/EEC Habitats directive). Plant Sociol. 2020, 57, 55–64. [Google Scholar] [CrossRef]
- Defeo, O.; McLachlan, A.; Schoeman, D.S.; Schlacher, T.A.; Dugan, J.; Jones, A.; Lastra, M.; Scapini, F. Threats to sandy beach ecosystems: A review. Estuar. Coast. Shelf Sci. 2009, 8, 1–12. [Google Scholar] [CrossRef]
- Chytrý, M.; Maskell, L.C.; Pino, J.; Pyšek, P.; Vilà, M.; Font, X.; Smart, S.M. Habitat invasons by alien plants: A quantitative comparison among Mediterranean, subcontinental and oceanic regions of Europe. J. Appl. Ecol. 2008, 45, 448–458. [Google Scholar] [CrossRef]
- Lazzaro, L.; Bolpagni, R.; Buffa, G.; Gentili, R.; Lonati, M.; Stinca, A.; Acosta, A.T.R.; Adorni, M.; Aleffi, M.; Allegrezza, M.; et al. Impact of invasive alien plants on native plant communities and Natura 2000 habitats: State of the art, gap analysis and perspectives in Italy. J. Environ. Manag. 2020, 274, 111140. [Google Scholar] [CrossRef] [PubMed]
- Tordoni, E.; Bacaro, G.; Weigelt, P.; Cameletti, M.; Janssen, J.A.M.; Acosta, A.T.R.; Bagella, S.; Filigheddu, R.; Bergmeier, E.; Buckley, H.L.; et al. Disentangling native and alien plant diversity in coastal sand dune ecosystems worldwide. J. Veg. Sci. 2020, 32, e12861. [Google Scholar] [CrossRef]
- Giulio, S.; Acosta, A.T.R.; Carboni, M.; Campos, J.A.; Chytrý, M.; Loidi, J.; Pergl, J.; Pyšek, P.; Isermann, M.; Janssen, J.A.M.; et al. Alien flora across European coastal dunes. Appl. Veg. Sci. 2020, 23, 317–327. [Google Scholar] [CrossRef]
- Royimani, L.; Mutanga, O.; Odindi, J.; Dube, T.; Matonger, T.N. Advancements in satellite remote sensing for mapping and monitoring of alien invasive plant species (AIPs). Phys. Chem. Earth PT A/B/C 2019, 112, 237–245. [Google Scholar] [CrossRef]
- Early, R.; Bradley, B.A.; Dukes, J.S.; Lawler, J.J.; Olden, J.D.; Blumenthal, D.M.; Gonzalez, P.; Grosholz, E.D.; Ibañez, I.; Miller, L.P.; et al. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 2016, 7, 12485. [Google Scholar] [CrossRef] [PubMed]
- Lambdon, P.W.; Pyšek, P.; Basnou, C.; Hejda, M.; Arianoutsou, M.; Essl, F.; Jarošík, V.; Pergl, J.; Winter, M.; Anastasiu, P.; et al. Alien flora of Europe: Species diversity, temporal trends, geographical patterns and research needs. Preslia 2008, 80, 101–149. [Google Scholar]
- Niphadkar, M.; Nagendra, H. Remote sensing of invasive plants: Incorporating functional traits into the picture. Int. J. Remote Sens. 2016, 37, 3074–3085. [Google Scholar] [CrossRef]
- Guisan, A.; Petitpierre, B.; Broennimann, O.; Daehler, C.; Kueffer, C. Unifying niche shift studies: Insights from biological invasions. Trends Ecol. Evol. 2014, 29, 260–269. [Google Scholar] [CrossRef] [Green Version]
- Drius, M.; Jones, L.; Marzialetti, F.; de Francesco, M.C.; Stanisci, A.; Carranza, M.L. Not just a sandy beach. The multi-service value of Mediterranean coastal dunes. Sci. Total Environ. 2019, 668, 1139–1155. [Google Scholar] [CrossRef] [Green Version]
- Viciani, D.; Vidali, M.; Gigante, D.; Bolpagni, R.; Villani, M.; Acosta, A.T.R.; Adorni, M.; Aleffi, M.; Allegrezza, M.; Angiolini, C.; et al. A first checklist of the alien-dominated vegetation in Italy. Plant Sociol. 2020, 57, 29–54. [Google Scholar] [CrossRef]
- Carranza, M.L.; Acosta, A.T.R.; Stanisci, A.; Pirone, G. Ecosystem classification for EU habitat distribution assessment in sandy coastal environments: An application in central Italy. Environ. Monit. Assess. 2008, 140, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Acosta, A.T.R.; Carranza, M.L.; Izzi, C.F. Are there habitats that contribute best to plant species diversity in coastal dunes? Biodivers. Conserv. 2009, 18, 1087–1098. [Google Scholar] [CrossRef]
- Sperandii, M.G.; Barták, V.; Acosta, A.T.R. Effectiveness of the Natura 2000 network in conserving Mediterranean coastal dune habitats. Biol. Conserv. 2020, 248, 108689. [Google Scholar] [CrossRef]
- Malavasi, M.; Santoro, R.; Cutini, M.; Acosta, A.T.R.; Carranza, M.L. The impact of human pressure on landscape patterns and plant species richness in Mediterranean coastal dunes. Plant Biosyst. 2016, 150, 73–82. [Google Scholar] [CrossRef]
- Campoy, J.G.; Acosta, A.T.R.; Affre, L.; Barreiro, R.; Brundu, G.; Buisson, E.; González, L.; Buisson, E.; González, L.; Lema, M. Monographs of invasive plants in Europe: Carpobrotus. Bot. Lett. 2018, 165, 440–475. [Google Scholar] [CrossRef]
- Sarmati, S.; Conti, L.; Acosta, A.T.R. Carpobrotus acinaciformis vs Carpobrotus edulis: Are there any differences in their impact on coastal dune plant biodiversity? Flora 2019, 257, 151422. [Google Scholar] [CrossRef]
- Carranza, M.L.; Carboni, M.; Feola, S.; Acosta, A.T.R. Landscape-scale patterns of alien plant species on coastal dune: The case of iceplant in central Italy. Appl. Veg. Sci. 2010, 13, 135–145. [Google Scholar] [CrossRef]
- Bazzichetto, M.; Malavasi, M.; Barták, V.; Acosta, A.T.R.; Moudrý, V.; Carranza, M.L. Modeling plant invasion on Mediterranean coastal landscapes: An integrative approach using remotely sensed data. Landsc. Urban Plan. 2018, 171, 98–106. [Google Scholar] [CrossRef]
- Santoro, R.; Jucker, T.; Carranza, M.L.; Acosta, A.T.R. Assessing the effects of Carpobrotus invasion on coastal dune soils. Does the nature of the invaded habitat matter? Community Ecol. 2011, 12, 234–240. [Google Scholar] [CrossRef]
- Conser, C.; Connor, E.F. Assessing the residual effects of Carpobrotus edulis invasion, implications for restoration. Biol. Invasions 2009, 11, 349–358. [Google Scholar] [CrossRef]
- Underwood, E.; Ustin, S.L.; Di Pietro, D. Mapping nonnative plants using hyperspectral imagery. Remote Sens. Environ. 2003, 86, 150–161. [Google Scholar] [CrossRef]
- Underwood, E.; Ustin, S.L.; Ramirez, C.M. A comparison of spatial and spectral image resolution for mapping invasive plants in coastal California. Environ. Manag. 2007, 39, 63–83. [Google Scholar] [CrossRef]
- Sperandii, M.G.; Prisco, I.; Stanisci, A.; Acosta, A.T.R. RanVegDunes-A random plot database of Italian coastal dunes. Phytocoenologia 2017, 47, 231–232. [Google Scholar] [CrossRef]
- Chytrý, M.; Tichý, L.; Hennekens, S.M.; Knollová, I.; Janssen, J.A.M.; Rodwell, J.S.; Peterka, T.; Marcenò, C.; Landucci, F.; Danihelka, J.; et al. EUNIS habitat classification: Expert system, characteristic species combinations and distribution maps of European habitats. Appl. Veg. Sci. 2020, 23, 1–28. [Google Scholar] [CrossRef]
- Cooley, S.W.; Smith, L.C.; Stepan, L.; Mascaro, J. Tracking dynamic northern surface water changes with high-frequency Planet CubeSat imagery. Remote Sens. 2017, 9, 1306. [Google Scholar] [CrossRef] [Green Version]
- Marzialetti, F.; Di Febbraro, M.; Malavasi, M.; Giulio, S.; Acosta, A.T.R.; Carranza, M.L. Mapping coastal dune landscape through spectral Rao’s Q temporal diversity. Remote Sens. 2020, 12, 2315. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S.A. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 118–126. [Google Scholar] [CrossRef]
- Wu, Z.; Velasco, M.; McVay, J.; Middleton, B.; Vogel, J.; Dye, D. MODIS derived vegetation index for drought detection on the San Carlos Apache reservation. Int. J. Adv. Remote Sens. GIS 2016, 5, 1524–1538. [Google Scholar] [CrossRef] [Green Version]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 55–107. [Google Scholar] [CrossRef]
- Mathieu, R.; Pouget, M.; Cervelle, B.; Escadafal, R. Relationships between satellite-based radiometric indices simulated using laboratory reflectance data and typic soil colour of an arid environment. Remote Sens. Environ. 1998, 66, 17–28. [Google Scholar] [CrossRef]
- Bachaoui, B.; Bachaoui, E.M.; Maimouni, S.; Lhissou, R.; El Harti, A.; El Ghmari, A. The use of spectral and geomorphometric data for water erosion mapping in El Ksiba region in the central High Atlas Mountains of Morocco. Appl. Geomat. 2014, 6, 159–169. [Google Scholar] [CrossRef]
- Escadafal, R. Remote sensing of soil color: Principles and applications. Remote Sens. Rev. 1993, 7, 261–279. [Google Scholar] [CrossRef]
- Whitaker, R.H. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Whitaker, R.H. Evolution and measurement of species diversity. Taxon 1972, 21, 213–251. [Google Scholar] [CrossRef] [Green Version]
- Magurran, A.E. Measuring Biological Diversity, 1st ed.; Blackwell Science: Maden, NJ, USA, 2004; pp. 100–185. [Google Scholar]
- Kindt, R.; Coe, R. Tree Diversity Analysis. A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies, 1st ed.; World Agroforestry Centre (ICRAF): Nairobi, Kenya, 2005; pp. 31–70. [Google Scholar]
- Fisher, R.A.; Corbet, A.S.; Williams, C.B. The relation between the number of species and the number of individuals in a random sample of an animal population. J. Anim. Ecol. 1943, 12, 42–58. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 23, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Oldeland, J.; Wesuls, D.; Rocchini, D.; Schmidt, M.; Jürgens, N. Does using species abundance data improve estimates of species diversity from remotely sensed spectral heterogeneity? Ecol. Indic. 2010, 10, 390–396. [Google Scholar] [CrossRef]
- Peet, R.K. The measurement of species diversity. Annu. Rev. Ecol. Syst. 1974, 5, 285–307. [Google Scholar] [CrossRef]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef] [Green Version]
- Borchers, H.W. Pracma: Pratical Numerical Math Functions, R Package Version 2.2.9. 2019. Available online: https://cran.r-project.org/web/packages/pracma/index.html (accessed on 15 October 2020).
- Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et du Jura. Bull. de la Société Vaud. des Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Wildi, O. Data Analysis in Vegetation Ecology, 1st ed.; John Wiley & Sons: Chichester, UK, 2010; pp. 25–34. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Pavoine, S.; Ricotta, C. Measuring functional dissimilarity among plots: Adapting old methods to new questions. Ecol. Indic. 2019, 97, 67–72. [Google Scholar] [CrossRef]
- Anderson, M.J.; Robinson, J. Permutation Tests for Linear Models. Aust. N. Z. J. Stat. 2001, 43, 75–88. [Google Scholar] [CrossRef]
- Frossard, J.; Renaud, O. Permuco: Permutation Tests for Regression, (Repeated Measures) ANOVA/ANCOVA and Comparison of Signals, R Package Version 1.1.0. 2019. Available online: https://cran.r-project.org/web/packages/permuco/index.html (accessed on 23 November 2020).
- Cade, B.S.; Noon, B.R. A gentle introduction to quantile regression for ecologists. Front. Ecol. Environ. 2003, 1, 412–420. [Google Scholar] [CrossRef]
- Henning, S.K.; Estrup, H.A.; Kathrin, K. Rejecting the mean: Estimating the response of fen plant species to environmental factors by non-linear quantile regression. J. Veg. Sci. 2005, 16, 373–382. [Google Scholar] [CrossRef]
- Koenker, R. Quantreg: Quantile Regression, R Package Version 5.73. 2020. Available online: https://cran.r-project.org/web/packages/quantreg/index.html (accessed on 10 November 2020).
- Warren, S.D.; Alt, M.; Olson, K.D.; Irl, S.D.H.; Steinbauer, M.J.; Jentsch, A. The relathionship between the spectral diversity of satellite imagery habitat heterogeneity, and plant species richness. Ecol. Inform. 2014, 24, 160–168. [Google Scholar] [CrossRef]
- Meng, J.; Li, S.; Wang, W.; Liu, Q.; Xie, S.; Ma, W. Estimation of forest structural diversity using the spectral and textural information derived from SPOT-5 satellite images. Remote Sens. 2016, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Arekhi, M.; Yilmaz, O.Y.; Yilmaz, H.; Akyüz, Y.F. Can tree species diversity be assessed with Landsat data in a temperate forest? Environ. Monit. Assess. 2017, 189, 586. [Google Scholar] [CrossRef] [PubMed]
- Rocchini, D.; McGlinn, D.; Ricotta, C.; Neteler, M.; Wohlgemuth, T. Landscape complexity and spatial scale influence the relathionship between remotely sensed spectral diversity and survey-based plant species richness. J. Veg. Sci. 2011, 22, 688–698. [Google Scholar] [CrossRef]
- Draper, F.C.; Baraloto, C.; Brodrick, P.G.; Phillips, O.L.; Martinez, R.V.; Honorio Coronado, E.N.; Baker, T.R.; Zárate Gómez, R.; Amasifuen Guerra, C.A.; Flores, M.; et al. Imaging spectroscopy predicts variable distance decay across contrasting Amazonian tree communities. J. Ecol. 2018, 107, 696–710. [Google Scholar] [CrossRef]
- Xu, Y.; Dickson, B.G.; Hampton, H.M.; Sisk, T.D.; Palumbo, J.A.; Prather, J.W. Effects of mismatches of scale and location between predictor and response variables on forest structure mapping. Photogramm. Engin. Remote Sens. 2009, 75, 313–322. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A.; Cavender-Bares, V.; Townsend, P.A.; Zygielbaum, A.I. The spatial sensitivity of the spectral diversity-biodiversity relationship: An experimental test in a prairie grassland. Ecol. Appl. 2018, 28, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Acosta, A.T.R.; Ercole, V.; Stanisci, A.; De Patta Pillar, V.; Blasi, C. Coastal vegetation zonation and dune morphology in some Mediterranean ecosystems. J. Coast. Res. 2007, 23, 1518–1524. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Stefanoni, J.L.; Gallardo-Cruz, J.A.; Meave, J.A.; Rocchini, D.; Bello-Pineda, J.; López-Martínez, J.O. Modeling α- and β-diversity in a tropical forest from remotely sensed and spatial data. Int. J. Appl. Earth Obs. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Meireles, J.E.; Cavender-Bares, J.; Townsend, P.A.; Ustin, S.; Gamon, J.A.; Schweiger, A.K.; Schaepman, M.E.; Asner, G.P.; Martin, R.E.; Singh, A.; et al. Leaf reflectance spectra capture the evolutionary history of seed plants. New Phytol. 2020, 228, 485–493. [Google Scholar] [CrossRef]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef]




| Acronym/Vegetation Categories | Description and Correspondence with EU Habitats (Ex Annex I 92/43/EEC) | Number of Plots |
|---|---|---|
| Shifting dunes/EUNIS-N14 | Mobile coastal sand ridges including embryonic dunes characterized by Elymus farctus and semi-permanent dune systems dominated by Ammophila arenaria subsp. australis (EU habitat code: 2110, 2120). | 79 |
| Transition dunes/EUNIS-N16 | Fixed dune grasslands including chamaephytic communities of the inland dunes dominated by Crucianella maritima and annual species-rich communities colonizing dry interdunal depressions. (EU habitat code: 2210, 2230). | 41 |
| Invaded dunes | Herbaceous vegetation with the presence Carpobrotus spp. covering more than 25 percent. | 43 |
| Acronym | Name | Bandwidth/Equation | Index of | Reference |
|---|---|---|---|---|
| B | Blue band | 455–515 nm | – | [61] |
| G | Green band | 500–590 nm | – | [61] |
| R | Red band | 590–670 nm | – | [61] |
| NIR | Near Infrared band | 780–860 nm | – | [61] |
| MSAVI2 | Modified Soil Adjusted Vegetation Index 2 | photosynthetic biomass | [63] | |
| CI | Colouration index | organic content level in soil | [66] |
| Acronym | Name | Formula | References |
|---|---|---|---|
| S | Species Richness | [73] | |
| H’ | Shannon index | [74] | |
| D | Inverse Simpson index | [76] |
| Acronym | Name | Formula | References |
|---|---|---|---|
| J | Jaccard similarity index | [79] | |
| BC | Bray-Curtis similarity index | [81] |
| Species Richness—Distance to Centroid | R2 = 0.383 | ||
| Linear Model with Interactions | Estimate | Std. Error | p-Value |
| Distance to centroid: Shifting dunes | 3.937 × 10−3 | 9.512 × 10−4 | 5.362 × 10−5 *** |
| Distance to centroid: Transition dunes | 4.554 × 10−3 | 8.794 × 10−4 | 6.666 × 10−7 *** |
| Distance to centroid: Invaded dunes | 4.746 × 10−3 | 8.903 × 10−4 | 3.311 × 10−7 *** |
| Shannon Index—Distance to Centroid | R2 = 0.322 | ||
| Linear Model with Interactions | Estimate | Std. Error | p-Value |
| Distance to centroid: Shifting dunes | 4.202 × 10−4 | 1.754 × 10−4 | 0.018 * |
| Distance to centroid: Transition dunes | 5.556 × 10−4 | 1.622 × 10−4 | 7.697 × 10−4 *** |
| Distance to centroid: Invaded dunes | 5.702× 10−4 | 1.642 × 10−4 | 6.624 × 10−4 *** |
| Inverse Simpson Index—Distance to Centroid | R2 = 0.342 | ||
| Linear Model with Interactions | Estimate | Std. Error | p-value |
| Distance to centroid: Shifting dunes | 1.695 × 10−3 | 6.450 × 10−4 | 9.426 × 10−3 ** |
| Distance to centroid: Transition dunes | 2.231 × 10−3 | 5.963 × 10−4 | 2.547 × 10−4 *** |
| Distance to centroid: Invaded dunes | 2.239 × 10−3 | 6.038 × 10−4 | 2.885 × 10−4 *** |
| Jaccard Similarities—Uclidean Distance | |||||
| Regression Type | τ | Intercept | Intercept Boundaries (99%) | Decay Rate (10−2) | Decay Rate (10−2) Boundaries (99%) |
| Linea regression | – | 0.145 *** | 0.138–0.152 | −1.207 *** | −1.405–−1.009 |
| Quantile regressions | 0.75 | 0.211 *** | 0.202–0.220 | −1.665 *** | −1.892–−1.412 |
| 0.90 | 0.318 *** | 0.306–0.330 | −2.312 *** | −2.655–−2.009 | |
| 0.95 | 0.377 *** | 0.361–0.393 | −2.426 *** | −2.83–−1.927 | |
| 0.99 | 0.521 *** | 0.491–0.564 | −2.226 *** | −3.208–−0.849 | |
| Bray-Curtis Similarities—Euclidean Distance | |||||
| Regression Type | τ | Intercept | Intercept Boundaries (99%) | Decay Rate (10−2) | Decay Rate (10−2) Boundaries (99%) |
| Linear regression | – | 0.235 *** | 0.207–0.257 | −1.814 *** | −2.106–−1.522 |
| Quantile | 0.75 | 0.353 *** | 0.339–0.367 | −2.529 *** | −2.867–−2.200 |
| 0.90 | 0.489 *** | 0.473–0.505 | −3.082 *** | −3.529–−2.614 | |
| 0.95 | 0.552 *** | 0.533–0.573 | −2.938 *** | −3.581–−2.206 | |
| 0.99 | 0.686 *** | 0.659–0.726 | −2.030 *** | −3.123–−0.699 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzialetti, F.; Cascone, S.; Frate, L.; Di Febbraro, M.; Acosta, A.T.R.; Carranza, M.L. Measuring Alpha and Beta Diversity by Field and Remote-Sensing Data: A Challenge for Coastal Dunes Biodiversity Monitoring. Remote Sens. 2021, 13, 1928. https://doi.org/10.3390/rs13101928
Marzialetti F, Cascone S, Frate L, Di Febbraro M, Acosta ATR, Carranza ML. Measuring Alpha and Beta Diversity by Field and Remote-Sensing Data: A Challenge for Coastal Dunes Biodiversity Monitoring. Remote Sensing. 2021; 13(10):1928. https://doi.org/10.3390/rs13101928
Chicago/Turabian StyleMarzialetti, Flavio, Silvia Cascone, Ludovico Frate, Mirko Di Febbraro, Alicia Teresa Rosario Acosta, and Maria Laura Carranza. 2021. "Measuring Alpha and Beta Diversity by Field and Remote-Sensing Data: A Challenge for Coastal Dunes Biodiversity Monitoring" Remote Sensing 13, no. 10: 1928. https://doi.org/10.3390/rs13101928