Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Area
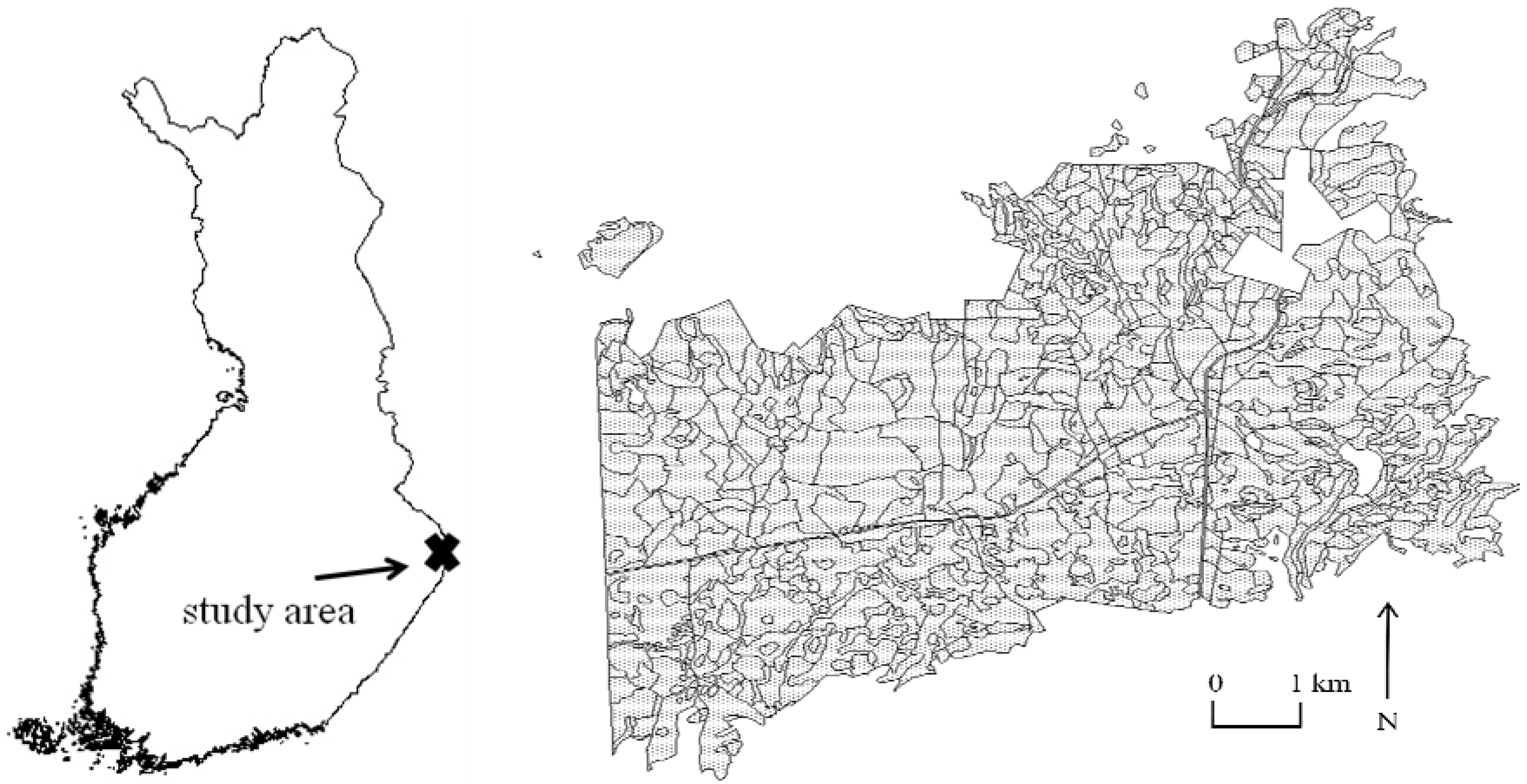
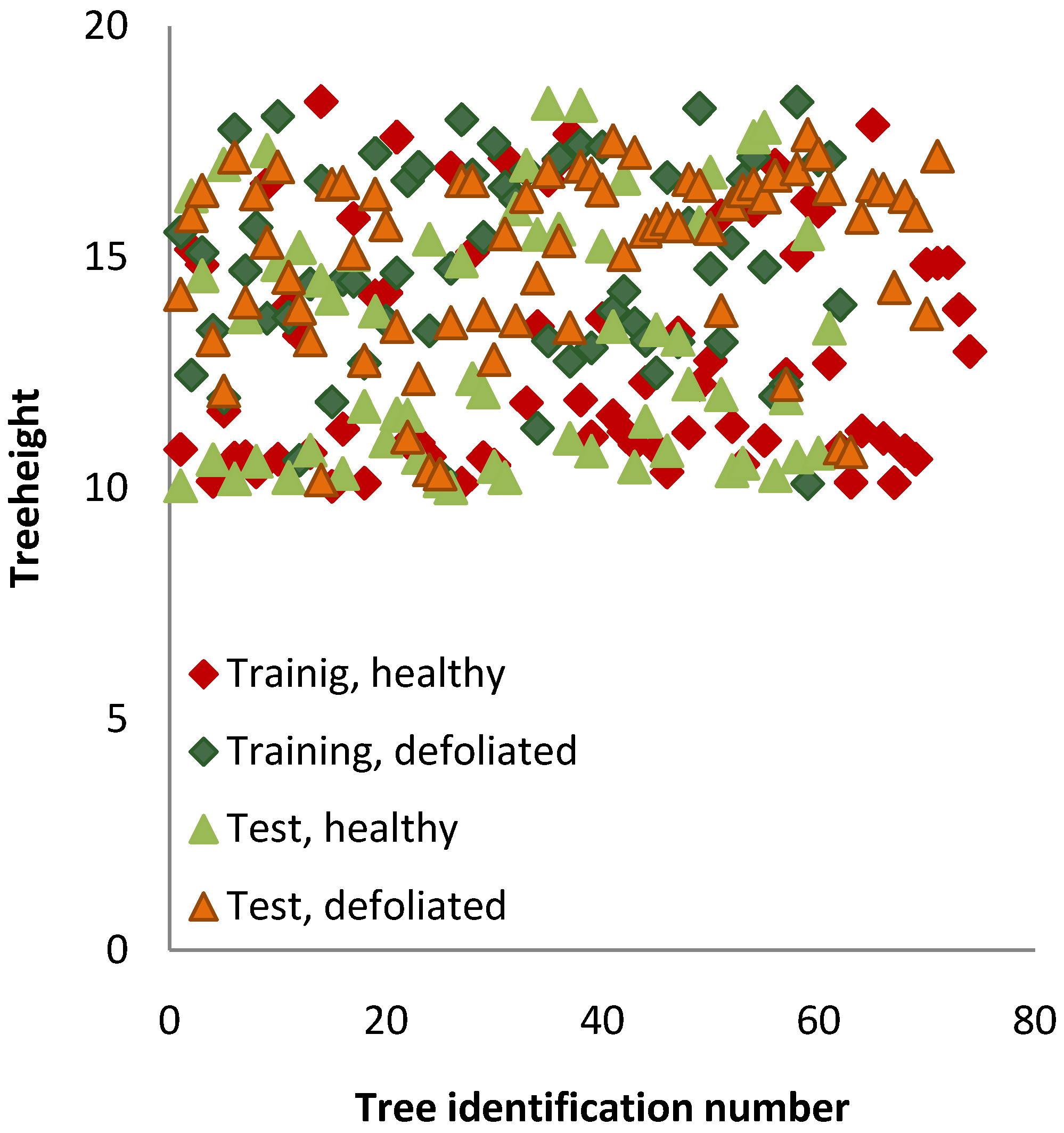
2.2. Field Data
| n | dmean | dmin | dmax | dsd | hmean | hmin | hmax | hsd | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Defoliated | 62 | 19.3 | 12.8 | 24.2 | 3.0 | 14.8 | 10.1 | 18.4 | 2.2 |
| Training | Healthy | 74 | 16.7 | 12.7 | 24.2 | 3.5 | 13.0 | 10.0 | 18.4 | 2.5 |
| Test | Defoliated | 74 | 19.7 | 12.9 | 23.2 | 2.7 | 15.1 | 10.2 | 17.7 | 2.0 |
| Test | Healthy | 61 | 17.0 | 12.6 | 24.2 | 3.6 | 13.2 | 10.0 | 18.3 | 2.6 |
2.3. Remote Sensing Material
2.4. Tree Detection and Feature Extraction
- (1).
- The CHM was smoothed with a Gaussian filter to remove small variations on the crown surface. The degree of smoothness was determined by the value of standard deviation (Gaussian scale) and kernel size of the filter.
- (2).
- Minimum curvatures were calculated. Minimum curvature is one of the principal curvatures. For a surface like CHM, a higher value of minimum curvature describes the tree top.
- (3).
- The smoothed CHM image was then scaled based on the computed minimum curvature resulting in a smoothed yet contrast-stretched image.
- (4).
- Local maxima were then searched for in a given neighborhood. These were considered as tree tops and used as markers in the following marker-controlled watershed transformation for tree crown delineations.
| Feature | Description |
|---|---|
| Hmax | Maximum laser height |
| Hmean | Arithmetic mean of laser heights |
| Hstd | Standard deviation of heights |
| CH | Crown height |
| CA | Crown area as a convex hull |
| CV | Crown volume as a convex hull in 3D |
| HP10-90 | Heights 0th–90th percentile |
| DS10-90 | Percentage of returns below 10–90% of total height |
| MaxD | Maximum crown diameter when crown was considered as an ellipse |
| Ponly | Percentage of only returns |
| Pfirst | Percentage of first returns |
| Pintermedian | Percentage of intermedian returns |
| Plast | Percentage of last returns |
| NIR | Spectral value of NIR band from aerial image |
| G | Spectral value of green band from aerial image |
| R | Spectral value of red band from aerial image |
2.5. Classification Methods
2.5.1. Logistic LASSO Regression
2.5.2. RF
2.5.3. MSN
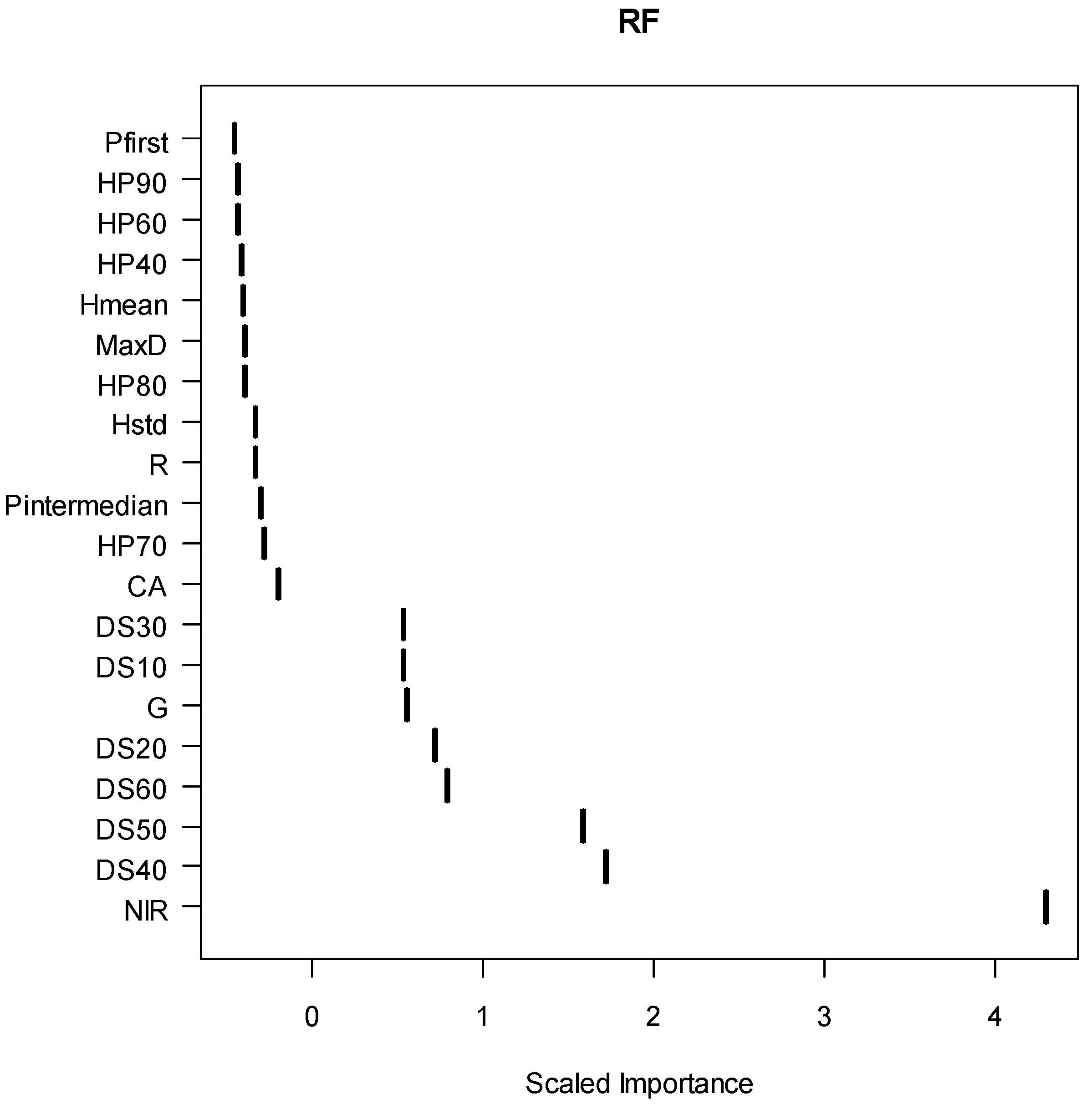
3. Results
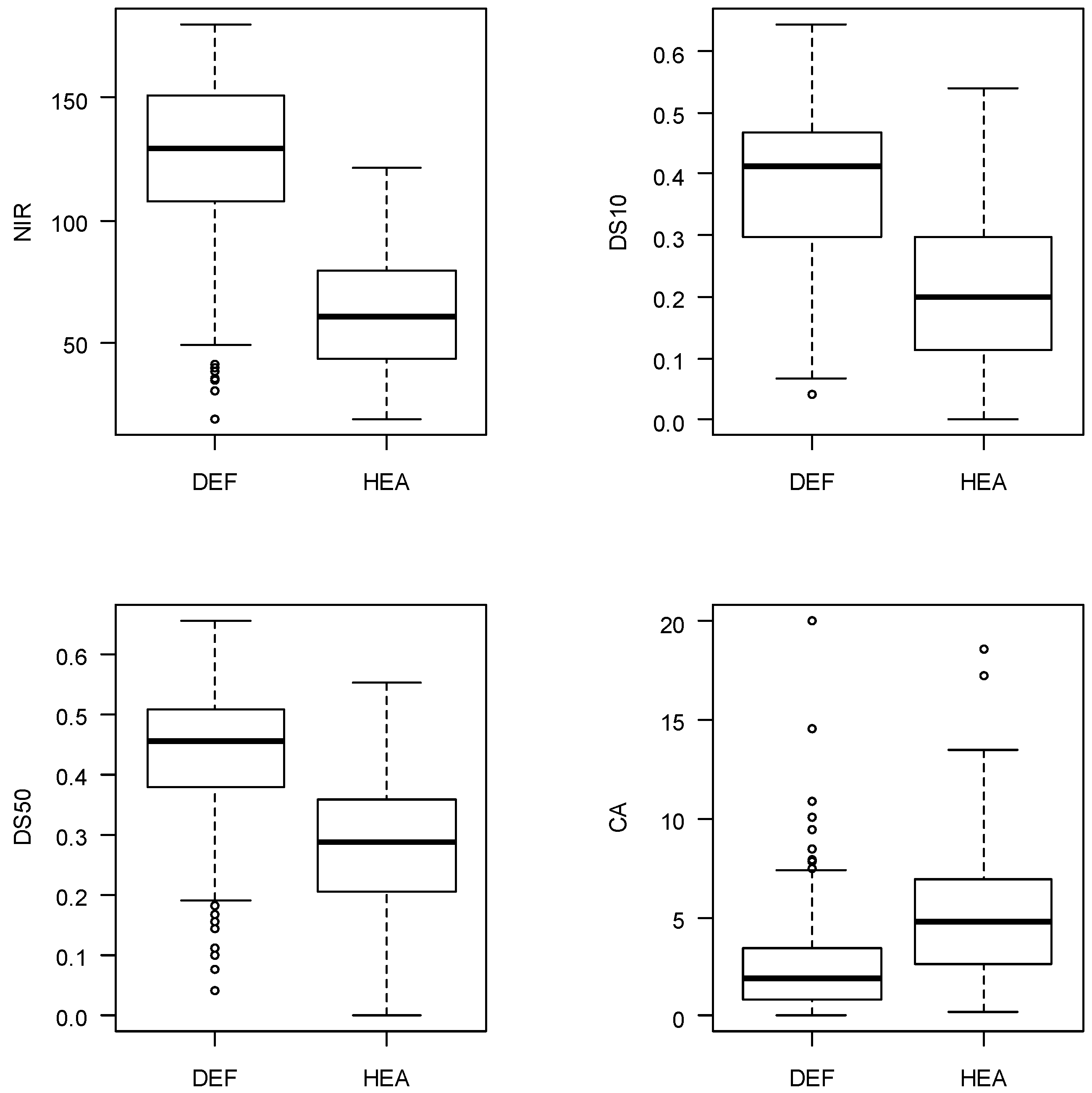
| Predictor | LASSO | RF | MSN | ||
|---|---|---|---|---|---|
| Estimate | |||||
| intercept | 5.6742999 | ||||
| CA | 0.1391846 | • | |||
| HP70 | • | ||||
| DS10 | −2.375357 | • | |||
| DS50 | −4.244455 | ||||
| DS60 | • | ||||
| NIR | −0.042489 | • | |||
| G | • | ||||
| Train set | Classification accuracy | 91.20% | 96.30% | 85.30% | |
| Kappa value | 0.81 | 0.91 | 0.66 | ||
| Test set | Classification accuracy | 86.7% | 88.1% | 83.7% | |
| Kappa value | 0.73 | 0.76 | 0.67 | ||
4. Discussion
5. Conclusions
Acknowledgements
References
- IPCC 2007. Summary for policymakers. In Climate Change 2007: The Physical Science Basis; Contribution of Working Group 1 to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Avyret, K.B., Tignor, M., Miller, H.L., Eds.; Cambridge University Press: Cambridge, UK and New York, NY, USA, 2007. [Google Scholar]
- Dale, V.H.; Joyce, L.A.; McNulty, S.; Neilson, R.P.; Ayres, M.P.; Flannican, M.D.; Hanson, P.J.; Irland, L.C.; Lugo, A.E.; Peterson, C.J.; Simberloff, D.; Swanson, F.J.; Stocks, B.J.; Wotton, B.M. Climate change and forest disturbances. Biosciences 2001, 51, 723–734. [Google Scholar] [CrossRef]
- Evans, H.; Straw, N.; Watt, A. Climate change: Implication for insect pests. In Climate Change: Impact on UK Forests; Roadmeadow, M., Ed.; Forestry Commission Bulletin 125; Forestry Commisssion: Edinburgh, UK, 2002; pp. 99–118. [Google Scholar]
- Walther, G.R.; Post, E.; Convey, P.; Menzel, A.; Parmesan, C.; Beebee, T.J.C.; Fromentin, J.-M.; Hoegh-Guldberg, O.; Bairlein, F. Ecological responses to recent climate change. Nature 2002, 416, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Logan, J.A.; Regniere, J.; Powell, J.A. Assessing the impacts of global warming on forest pest dynamics. Front. Ecol. Environ. 2003, 1, 130–137. [Google Scholar] [CrossRef]
- Lyytikäinen-Saarenmaa, P.; Tomppo, E. Impact of sawfly defoliation on growth of Scots pine Pinus sylvestris (Pinaceae) and associated economic losses. Bull. Entomol. Res. 2002, 92, 137–140. [Google Scholar] [CrossRef] [PubMed]
- De Somviele, B.; Lyytikäinen-Saarenmaa, P.; Niemelä, P. Stand edge effects on distribution and condition of Diprionid sawflies. Agr. Forest Entomol. 2007, 9, 17–30. [Google Scholar] [CrossRef]
- Lyytikäinen-Saarenmaa, P.; Niemelä, P.; Annila, E. Growth responses and mortality of Scots pine (Pinus sylvestris L.) after a pine sawfly outbreak. In Forest Insect Population Dynamics and Host Influences, In Proceedings of International Symposium of IUFRO, Kanazawa, Japan, September 14–19, 2003; Kamata, N., Liebhold, A.L., Quiring, D.T., Clancy, K.M., Eds.; Kanazawa University: Kanazawa, Ishikawa, Japan, 2003; pp. 81–85. [Google Scholar]
- Viitasaari, M.; Varama, M. Sawflies 4. Conifer sawflies (Diprionidae); Department of Agricultural and Forest Zoology Reports; University of Helsinki: Helsinki, Finland, 1987; pp. 1–79, (In Finnish with an English summary). [Google Scholar]
- Geri, G. The pine sawfly in central France. In Dynamics of Forest Insect Populations: Patterns, Causes, Implications; Berryman, A.A., Ed.; Plenum Press: New York, NY, USA, 1988; pp. 377–405. [Google Scholar]
- De Somviele, B.; Lyytikäinen-Saarenmaa, P.; Niemelä, P. Sawfly (Hym. Diprionidae) outbreaks on Scots pine: Effect of stand structure, site quality and relative tree position on defoliation intensity. Forest Ecol. Manag. 2004, 194, 305–317. [Google Scholar] [CrossRef]
- Lyytikäinen-Saarenmaa, P.; Holopainen, M.; Ilvesniemi, S.; Haapanen, R. Detecting pine sawfly defoliation by means of remote sensing and GIS. Forstschutz Aktuell. 2008, 44, 14–15. [Google Scholar]
- Ciesla, W. Remote Sensing in Forest Health Protection; USDA Forest Service Remote Sensing Applications Center: Salt Lake City; UT and Forest Health Technology Enterprise Team: Fort Collins, CO, USA, 2000.
- Hall, R.J.; Skakun, R.S.; Arsenault, E.J. Remotely sensed data in the mapping of insect defoliation. In Understanding Forest Disturbance and Spatial Pattern: Remote Sensing and GIS Approaches; Michael, W.A., Franklin, S.E., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2007; pp. 85–111. [Google Scholar]
- Falkowski, M.J.; Smith, A.M.S.; Hudak, A.T.; Gessler, P.E.; Vierling, L.A.; Crookston, N.L. Automated estimation of individual conifer tree height and crown diameter via two-dimensional spatial wavelet analysis of lidar data. Can. J. Remote Sens. 2006, 32, 153–161. [Google Scholar] [CrossRef]
- Hyyppä, J.; Inkinen, M. Detecting and estimating attributes for single trees using laser scanner. Photogramm. J. Fin. 1999, 16, 27–42. [Google Scholar]
- Magnussen, S.; Eggermont, P.; LaRiccia, V.N. Recovering tree heights from airborne laser scanner data. Forest Sci. 1999, 45, 407–422. [Google Scholar]
- Maltamo, M.; Mustonen, K.; Hyyppä, J.; Pitkänen, J.; Yu, X. The accuracy of estimating individual tree variables with airborne laser scanning in boreal nature reserves. Can. J. Forest Res. 2004, 34, 1791–1801. [Google Scholar] [CrossRef]
- Bortolot, Z.; Wynne, R.H. Estimating forest biomass using small footprint LiDAR data: An individual tree-based approach that incorporates training data. ISPRS J. Photogramm. Remote Sens. 2005, 59, 342–360. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.; Cohen, W.B.; Parker, G.; Shugart, H.H. Surface lidar remote sensing of basal area and biomass in deciduous forests of Eastern Maryland, USA. Remote Sens. Environ 1999, 67, 83–98. [Google Scholar] [CrossRef]
- Van Aardt, J.A.N.; Wynne, R.H.; Scrivani, J.A. Lidar-based mapping of forest volume and biomass by taxonomic group using structurally homogenous segments. Photogramm. Eng. Remote Sensing 2008, 74, 1033–1044. [Google Scholar] [CrossRef]
- Hyyppä, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Næsset, E. Estimating timber volume of forest stands using airborne laser scanner data. Remote Sens. Environ 1997, 61, 246–253. [Google Scholar] [CrossRef]
- Wallerman, J.; Holmgren, J. Estimating field-plot data of forest stands using airborne laser scanning and SPOT HRG data. Remote Sens. Environ. 2007, 110, 501–508. [Google Scholar] [CrossRef]
- Means, J.E.; Acker, S.A.; Fitt, B.J.; Renslow, M.; Emerson, L.; Hendrix, C.J. Predicting forest stand characteristics with airborne scanning lidar. Photogramm. Eng. Remote Sensing. 2000, 66, 1367–1371. [Google Scholar]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne lidar. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree species classification using airborne LiDAR—Effects of stand and tree parameters, downsizing of training set, intensity normalization, and sensor type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Korpela, I.; Maltamo, M.; Tokola, T. Imputation of single-tree attributes using airborne laser scanning-based height, intensity and alpha shape metrics. Remote Sens. Environ. 2010, 114, 1263–1276. [Google Scholar] [CrossRef]
- Sohlberg, S.; Næsset, E.; Hanssen, K.H.; Christiansen, E. Mapping defoliation during a severe insect attack on Scots pine using airborne laser scanning. Remote Sens. Environ. 2006, 102, 364–376. [Google Scholar] [CrossRef]
- Solberg, S.; Næsset, E. Monitoring forest health by remote sensing. In Symposium on Forests in a Changing Environment—Results of 20 years ICP Forests Monitoring, Göttingen, Germany, October 25–28, 2006; Schriftenreihe der Forstlichen Fakultät Göttingen und der Nordwestdeutschen Forstlichen Versuchsanstalt: Göttingen, Germany, 2007a; pp. 99–104. [Google Scholar]
- Solberg, S. Mapping gap fraction, LAI and defoliation using various ALS penetration variables. Int. J. Remote Sens. 2010, 32, 1227–1244. [Google Scholar] [CrossRef]
- Solberg, S.; Næsset, E. Mapping defoliation with LIDAR. In ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, September 12–14, 2007; Volume 3/W52, pp. 379–382.
- Eichhorn, J. Manual on Methods and Criteria for Harmonized Sampling, Assessment, Monitoring and Analysis of the Effects of Air Pollution on Forests. Part II. Visual Assessment of Crown Condition and Submanual on Visual Assessment of Crown Condition on Intensive Monitoring Plots; United Nations Economic Commission for Europe Convention on Long-range Transboundary Air Pollution: Hamburg, Germany, 1998. [Google Scholar]
- Lyytikäinen-Saarenmaa, P. Unpublished data.
- Axelsson, P. DEM generation from laser scanner data using adaptive TIN models. In XIX ISPRS Congress, Commission I-VII, Amsterdam, The Netherlands, July 16–23, 2000; pp. 110–117.
- Yu, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on random forest technique. ISPRS J. Photogramm. Remote Sens. 2010. In press. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. Royal. Statist. Soc B. 1996, 58, 267–288. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2007. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Crookston, N.L.; Finley, A.O. A R package for efficient nearest neighbor imputation routines, variance estimation, and mapping, 2007-2010. Available online: http://cran.r-project.org (accessed on 10 July 2010).
- Moeur, M.; Stage, A.R. Most Similar Neighbor: An improved sampling inference procedure for natural resource planning. Forest Sci. 1995, 41, 337–359. [Google Scholar]
- Goldberg, D.E. Genetic Algorithms in Search, Optimization, and Machine Learning; Addison-Wesley: Boston, MA, USA, 1989; p. 372. [Google Scholar]
- Trevino, V.; Falciani, F. GALGO: A R package for multivariate variable selection using genetic algorithms. Bioinformatics 2006, 22, 1154–1156. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, M.; Haapanen, R.; Tuominen, S.; Viitala, R. Performance of airborne laser scanning- and aerial photograph-based statistical and textural features in forest variable estimation. In Proceedings of Silvilaser 2008, Edinburgh, UK, September 17–19, 2008; pp. 105–112.
- Packalén, P.; Maltamo, M. The k-MSN method in the prediction of species specific stand attributes using airborne laser scanning and aerial photographs. Remote Sens. Environ. 2007, 109, 328–341. [Google Scholar] [CrossRef]
- Vastaranta, M.; Holopainen, M.; Haapanen, R.; Yu, X.; Melkas, T.; Hyyppä, H.; Hyyppä, J. Individual tree detection, lasso regression and k-nearest neighbour methods in retrieval of forest inventory characteristics from low-pulse airborne laser scanning. 2010; Submitted. [Google Scholar]
- Tomppo, E. The Finnish National Forest Inventory. In Forest Inventory. Methodology and Applications (Managing Forest Ecosystems); Kangas, A., Maltamo, M., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 179–194. [Google Scholar]
- Ilvesniemi, S. Numeeriset ilmakuvat ja Landsat TM -satelliittikuvat männyn neulaskadon arvioinnissa; Pro Gardu. Metsävarojen käytön laitos, Helsingin yliopisto: Helsinki, Finland, 2009; p. 62. (In Finnish) [Google Scholar]
- Haara, A.; Nevalainen, S. Detection of dead or defoliated spruces using digital aerial data. Forest Ecol Manag. 2002, 160, 97–107. [Google Scholar] [CrossRef]
- Fukuda, K.; Pearson, P.A. Data mining and image segmentation approaches for classifying defoliation in aerial forest imagery. In Proceedings of 3rd Biennial Meeting of the IEMSs, Burlington, VT, USA, July 9–13, 2006.
- Karjalainen, M.; Kaasalainen, S.; Hyyppä, J.; Holopainen, M.; Lyytikäinen-Saarenmaa, P.; Krooks, A.; Jaakkola, A. SAR Satellite Images and Terrestrial Laser Scanning in Forest Damages Mapping in Finland. In Proceedings of ESA Living Planet Symposium, Bergen, Norway, June 28–July 2, 2010. SP-686.
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kantola, T.; Vastaranta, M.; Yu, X.; Lyytikainen-Saarenmaa, P.; Holopainen, M.; Talvitie, M.; Kaasalainen, S.; Solberg, S.; Hyyppa, J. Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images. Remote Sens. 2010, 2, 2665-2679. https://doi.org/10.3390/rs2122665
Kantola T, Vastaranta M, Yu X, Lyytikainen-Saarenmaa P, Holopainen M, Talvitie M, Kaasalainen S, Solberg S, Hyyppa J. Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images. Remote Sensing. 2010; 2(12):2665-2679. https://doi.org/10.3390/rs2122665
Chicago/Turabian StyleKantola, Tuula, Mikko Vastaranta, Xiaowei Yu, Paivi Lyytikainen-Saarenmaa, Markus Holopainen, Mervi Talvitie, Sanna Kaasalainen, Svein Solberg, and Juha Hyyppa. 2010. "Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images" Remote Sensing 2, no. 12: 2665-2679. https://doi.org/10.3390/rs2122665
APA StyleKantola, T., Vastaranta, M., Yu, X., Lyytikainen-Saarenmaa, P., Holopainen, M., Talvitie, M., Kaasalainen, S., Solberg, S., & Hyyppa, J. (2010). Classification of Defoliated Trees Using Tree-Level Airborne Laser Scanning Data Combined with Aerial Images. Remote Sensing, 2(12), 2665-2679. https://doi.org/10.3390/rs2122665








