An Optimal Sample Data Usage Strategy to Minimize Overfitting and Underfitting Effects in Regression Tree Models Based on Remotely-Sensed Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case Study Area
2.2. Data Used for Developing and Evaluating Regression Tree Models
2.2.1. Background
2.2.2. Landsat 8 Observations and MODIS NDVI Data
2.2.3. Spatial Evaluations of Large Model Prediction Errors Using NLCD 2011
2.3. Method and Procedures
2.3.1. Method
2.3.2. Processing Procedures
3. Results and Discussion
3.1. Identification of the Optimal Regression Tree Model Complexity and Associated Model Parameters
3.2. Model Accuracy Assessments for the Four Conditions
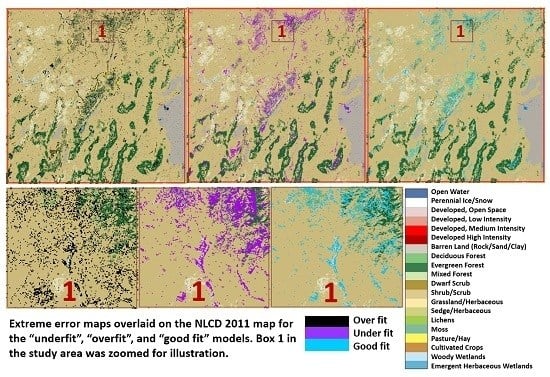
3.3. Evaluation of the Extreme AD Regions for the Four Models Using NLCD 2011
3.4. Discussion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Anderson, J.R.; Hardy, E.E.; Roach, J.T.; Witmer, R.E. A Land Use and Land Cover Classification System for Use with Remote Sensor Data; P 0964; U.S. Geological Survey: Reston, VA, USA, 1976; p. 28.
- Gu, Y.; Brown, J.F.; Miura, T.; van Leeuwen, W.J.; Reed, B.C. Phenological classification of the United States: A geographic framework for extending multi-sensor time-series data. Remote Sens. 2010, 2, 526–544. [Google Scholar] [CrossRef]
- Wylie, B.K.; Zhang, L.; Bliss, N.B.; Ji, L.; Tieszen, L.L.; Jolly, W.M. Integrating modelling and remote sensing to identify ecosystem performance anomalies in the boreal forest, Yukon River Basin, Alaska. Int. J. Digit. Earth 2008, 1, 196–220. [Google Scholar] [CrossRef]
- Gu, Y.; Wylie, B.K. Detecting ecosystem performance anomalies for land management in the upper colorado river basin using satellite observations, climate data, and ecosystem models. Remote Sens. 2010, 2, 1880–1891. [Google Scholar] [CrossRef]
- Homer, C.; Dewitz, J.; Yang, L.; Jin, S.; Danielson, P.; Xian, G.; Coulston, J.; Herold, N.; Wickham, J.; Megown, K. Completion of the 2011 national land cover database for the conterminous United States–representing a decade of land cover change information. Photogramm. Eng. Remote Sens. 2015, 81, 345–354. [Google Scholar]
- Homer, C.G.; Aldridge, C.L.; Meyer, D.K.; Schell, S.J. Multi-scale remote sensing sagebrush characterization with regression trees over wyoming, USA: Laying a foundation for monitoring. Int. J. Appl. Earth Obs. Geoinf. 2012, 14, 233–244. [Google Scholar] [CrossRef]
- Peters, A.J.; Walter-Shea, E.A.; Ji, L.; Viña, A.; Hayes, M.; Svoboda, M.D. Drought monitoring with ndvi-based standardized vegetation index. Photogramm. Eng. Remote Sens. 2002, 68, 71–75. [Google Scholar]
- Potter, C.S.; Randerson, J.T.; Field, C.B.; Matson, P.A.; Vitousek, P.M.; Mooney, H.A.; Klooster, S.A. Terrestrial ecosystem production: A process model based on global satellite and surface data. Glob. Biogeochem. Cycles 1993, 7, 811–841. [Google Scholar] [CrossRef]
- Tucker, C.J.; Vanpraet, C.L.; Sharman, M.J.; van Ittersum, G. Satellite remote sensing of total herbaceous biomass production in the senegalese sahel: 1980–1984. Remote Sens. Environ. 1985, 17, 233–249. [Google Scholar] [CrossRef]
- Reed, B.C.; Brown, J.F.; Vanderzee, D.; Loveland, T.R.; Merchant, J.W.; Ohlen, D.O. Measuring phenological variability from satellite imagery. J. Veg. Sci. 1994, 5, 703–714. [Google Scholar] [CrossRef]
- Loveland, T.R.; Reed, B.C.; Brown, J.F.; Ohlen, D.O.; Zhu, Z.; Yang, L.; Merchant, J.W. Development of a global land cover characteristics database and IGBP DISCover from 1 km AVHRR data. Int. J. Remote Sens. 2000, 21, 1303–1330. [Google Scholar] [CrossRef]
- Washington-Allen, R.A.; West, N.E.; Ramsey, R.D.; Efroymson, R.A. A protocol for retrospective remote sensing-based ecological monitoring of rangelands. Rangel. Ecol. Manag. 2006, 59, 19–29. [Google Scholar] [CrossRef]
- Burgan, R.E.; Klaver, R.W.; Klarer, J.M. Fuel models and fire potential from satellite and surface observations. Int. J. Wildland Fire 1998, 8, 159–170. [Google Scholar] [CrossRef]
- Zhu, Z.; Woodcock, C.E. Continuous change detection and classification of land cover using all available Landsat data. Remote Sens. Environ. 2014, 144, 152–171. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Liao, A.; Cao, X.; Chen, L.; Chen, X.; He, C.; Han, G.; Peng, S.; Lu, M.; et al. Global land cover mapping at 30 m resolution: A pok-based operational approach. ISPRS J. Photogramm. Remote Sens. 2015, 103, 7–27. [Google Scholar] [CrossRef]
- Giri, C.; Pengra, B.; Long, J.; Loveland, T.R. Next generation of global land cover characterization, mapping, and monitoring. Int. J. Appl. Earth Obs. Geoinform. 2013, 25, 30–37. [Google Scholar] [CrossRef]
- Reed, B.C.; White, M.A.; Brown, J.F. Remote sensing phenology. In Phenology: An Integrative Environmental Science; Schwartz, M.D., Ed.; Kluwer Academic Publ.: Dordrecht, The Netherlands, 2003; pp. 365–381. [Google Scholar]
- Tan, Z.; Liu, S.; Wylie, B.K.; Jenkerson, C.B.; Oeding, J.; Rover, J.; Young, C. MODIS-informed greenness responses to daytime land surface temperature fluctuations and wildfire disturbances in the Alaskan Yukon River Basin. Int. J. Remote Sens. 2012, 34, 2187–2199. [Google Scholar] [CrossRef]
- White, M.A.; de Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; van Leeuwen, W.J.D.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Becker-Reshef, I.; Vermote, E.; Lindeman, M.; Justice, C. A generalized regression-based model for forecasting winter wheat yields in Kansas and Ukraine using MODIS data. Remote Sens. Environ. 2010, 114, 1312–1323. [Google Scholar] [CrossRef]
- Howard, D.M.; Wylie, B.K.; Tieszen, L.L. Crop classification modelling using remote sensing and environmental data in the greater Platte River Basin, USA. Int. J. Remote Sens. 2012, 33. [Google Scholar] [CrossRef]
- Wylie, B.K.; Boyte, S.P.; Major, D.J. Ecosystem performance monitoring of rangelands by integrating modeling and remote sensing. Rangel. Ecol. Manag. 2012, 65. [Google Scholar] [CrossRef]
- Park, S.; Im, J.; Jang, E.; Rhee, J. Drought assessment and monitoring through blending of multi-sensor indices using machine learning approaches for different climate regions. Agric. Forest Meteorol. 2016, 216, 157–169. [Google Scholar] [CrossRef]
- Yang, F.; Ichii, K.; White, M.A.; Hashimoto, H.; Michaelis, A.R.; Votava, P.; Zhu, A.X.; Huete, A.; Running, S.W.; Nemani, R.R. Developing a continental-scale measure of gross primary production by combining MODIS and ameriflux data through support vector machine approach. Remote Sens. Environ. 2007, 110, 109–122. [Google Scholar] [CrossRef]
- Xiao, J.; Zhuang, Q.; Law, B.E.; Chen, J.; Baldocchi, D.D.; Cook, D.R.; Oren, R.; Richardson, A.D.; Wharton, S.; Ma, S.; et al. A continuous measure of gross primary production for the conterminous United States derived from MODIS and ameriflux data. Remote Sens. Environ. 2010, 114, 576–591. [Google Scholar] [CrossRef]
- Zhang, L.; Wylie, B.K.; Ji, L.; Gilmanov, T.G.; Tieszen, L.L.; Howard, D.M. Upscaling carbon fluxes over the great plains grasslands: Sinks and sources. J. Geophys. Res. Biogeosci. 2011, 116. [Google Scholar] [CrossRef]
- RuleQuest Research. Available online: http://www.rulequest.com/ (accessed on 10 November 2016).
- Zhang, L.; Wylie, B.K.; Ji, L.; Gilmanov, T.G.; Tieszen, L.L. Climate-driven interannual variability in net ecosystem exchange in the Northern Great Plains Grasslands. Rangel. Ecol. Manag. 2010, 63, 40–50. [Google Scholar] [CrossRef]
- Gu, Y.; Wylie, B. Downscaling 250-m MODIS growing season NDVI based on multiple-date Landsat images and data mining approaches. Remote Sens. 2015, 7, 3489–3506. [Google Scholar] [CrossRef]
- Boyte, S.P.; Wylie, B.K.; Major, D.J.; Brown, J.F. The integration of geophysical and enhanced moderate resolution imaging spectroradiometer normalized difference vegetation index data into a rule-based, piecewise regression-tree model to estimate cheatgrass beginning of spring growth. Int. J. Digit. Earth 2013, 8. [Google Scholar] [CrossRef]
- Brown, J.F.; Wardlow, B.D.; Tadesse, T.; Hayes, M.J.; Reed, B.C. The vegetation drought response index (vegdri): A new integrated approach for monitoring drought stress in vegetation. GISci. Remote Sens. 2008, 45, 16–46. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Smale, C. Best choices for regularization parameters in learning theory: On the bias—Variance problem. Found. Comput. Math. 2002, 2, 413–428. [Google Scholar]
- Yu, L.; Lai, K.K.; Wang, S.; Huang, W. A bias-variance-complexity trade-off framework for complex system modeling. In Computational Science and Its Applications—ICCSA 2006: International Conference, Glasgow, Uk, 8–11 May 2006. Proceedings, Part I; Gavrilova, M., Gervasi, O., Kumar, V., Tan, C.J.K., Taniar, D., Laganá, A., Mun, Y., Choo, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 518–527. [Google Scholar]
- Quinlan, J.R. Combining instance-based and model-based learning. In Proceedings of the Tenth International Conference on Machine Learning, Amherst, MA, USA, 27–29 June 1993; pp. 236–243.
- Rouse, J.W., Jr.; Haas, H.R.; Deering, D.W.; Schell, J.A.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NTRS: Greenbelt, MD, USA, 1974; p. 371. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Chen, D.; Brutsaert, W. Satellite-sensed distribution and spatial patterns of vegetation parameters over a Tallgrass Prairie. J. Atmos. Sci. 1998, 55, 1225–1238. [Google Scholar] [CrossRef]
- Funk, C.; Budde, M.E. Phenologically-tuned MODIS NDVI-based production anomaly estimates for Zimbabwe. Remote Sens. Environ. 2009, 113, 115–125. [Google Scholar] [CrossRef]
- Gu, Y.; Wylie, B.K.; Bliss, N.B. Mapping grassland productivity with 250-m emodis NDVI and ssurgo database over the greater Platte River Basin, USA. Ecol. Indic. 2013, 24, 31–36. [Google Scholar] [CrossRef]
- MODIS Products Table. Available online: https://lpdaac.usgs.gov/dataset_discovery/modis/modis_products_table (accessed on 10 November 2016).
- Tieszen, L.L.; Reed, B.C.; Bliss, N.B.; Wylie, B.K.; DeJong, D.D. NDVI, C3 and C4 production, and distributions in Great Plains grassland land cover classes. Ecol. Appl. 1997, 7, 59–78. [Google Scholar]
- Wylie, B.K.; Denda, I.; Pieper, R.D.; Harrington, J.A.; Reed, B.C.; Southward, G.M. Satellite-based herbaceous biomass estimates in the pastoral zone of Niger. J. Range Manag. 1995, 48, 159–164. [Google Scholar] [CrossRef]
- Gu, Y.; Wylie, B.K. Developing a 30-m grassland productivity estimation map for Central Nebraska using 250-m MODIS and 30-m Landsat-8 observations. Remote Sens. Environ. 2015, 171, 291–298. [Google Scholar] [CrossRef]
- Nelson, K.J.; Steinwand, D. A Landsat data tiling and compositing approach optimized for change detection in the conterminous United States. Photogramm. Eng. Remote Sens. 2015, 81, 573–586. [Google Scholar] [CrossRef]
- USGS eMODIS Data. Available online: https://lta.cr.usgs.gov/emodis (accessed on 10 November 2016).
- Jenkerson, C.B.; Maiersperger, T.K.; Schmidt, G.L. Emodis—A User-Friendly Data Source; U.S. Geological Survey Open-File Report 2010-1055; U.S. Geological Survey Earth Resources Observation and Science (EROS) Center: Sioux Falls, SD, USA, 2010.
- Swets, D.L.; Reed, B.C.; Rowland, J.R.; Marko, S.E. A weighted least-squares approach to temporal smoothing of NDVI. In Proceedings of the ASPRS Annual Conference, From Image to Information, Portland, Oregon, 17–21 May 1999.
- Brown, F.J.; Howard, D.; Wylie, B.; Frieze, A.; Ji, L.; Gacke, C. Application-ready expedited MODIS data for operational land surface monitoring of vegetation condition. Remote Sens. 2015, 7, 16226–16240. [Google Scholar] [CrossRef]
- National Land Cover Database 2011. Available online: http://www.mrlc.gov/nlcd2011.php (accessed on 10 November 2016).
- Python Software Foundation. Available online: https://www.python.org/ (accessed on 10 November 2016).
- Gu, Y.; Wylie, B.K.; Boyte, S.P. Landsat 8 Six Spectral Band Data and MODIS NDVI Data for Assessing the Optimal Regression Tree Models. Available online: https://dx.doi.org/10.5066/F7319T1P (accessed on 10 November 2016).
- Cawley, G.C.; Talbot, N.L.C. Fast exact leave-one-out cross-validation of sparse least-squares support vector machines. Neural Netw. 2004, 17, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Wylie, B.K.; Johnson, D.A.; Laca, E.A.; Saliendra, N.Z.; Gilmanov, T.G.; Reed, B.C.; Tieszen, L.L.; Worstell, B.B. Calibration of remotely sensed, coarse resolution NDVI to co 2 fluxes in a sagebrush-steppe ecosystem. Remote Sens. Environ. 2003, 85, 243–255. [Google Scholar] [CrossRef]
- Wylie, B.K.; Fosnight, E.A.; Gilmanov, T.G.; Frank, A.B.; Morgan, J.A.; Haferkamp, M.R.; Meyers, T.P. Adaptive data-driven models for estimating carbon fluxes in the Northern Great Plains. Remote Sens. Environ. 2007, 106, 399–413. [Google Scholar] [CrossRef]
- Ji, L.; Wylie, B.K.; Nossov, D.R.; Peterson, B.E.; Waldrop, M.P.; McFarland, J.W.; Rover, J.A.; Hollingsworth, T.N. Estimating aboveground biomass in interior alaska with Landsat data and field measurements. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 451–461. [Google Scholar] [CrossRef]
- Xiao, J.; Ollinger, S.V.; Frolking, S.; Hurtt, G.C.; Hollinger, D.Y.; Davis, K.J.; Pan, Y.; Zhang, X.; Deng, F.; Chen, J.; et al. Data-driven diagnostics of terrestrial carbon dynamics over North America. Agric. For. Meteorol. 2014, 197, 142–157. [Google Scholar] [CrossRef]







| Overfitting | Underfitting | Good Fit | |
|---|---|---|---|
| MAD | MADtraining < MADtesting | Highest MADtraining and highest MADtesting | Lowest MADtraining and lowest MADtesting |
| SD | High | High | Relatively low |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Wylie, B.K.; Boyte, S.P.; Picotte, J.; Howard, D.M.; Smith, K.; Nelson, K.J. An Optimal Sample Data Usage Strategy to Minimize Overfitting and Underfitting Effects in Regression Tree Models Based on Remotely-Sensed Data. Remote Sens. 2016, 8, 943. https://doi.org/10.3390/rs8110943
Gu Y, Wylie BK, Boyte SP, Picotte J, Howard DM, Smith K, Nelson KJ. An Optimal Sample Data Usage Strategy to Minimize Overfitting and Underfitting Effects in Regression Tree Models Based on Remotely-Sensed Data. Remote Sensing. 2016; 8(11):943. https://doi.org/10.3390/rs8110943
Chicago/Turabian StyleGu, Yingxin, Bruce K. Wylie, Stephen P. Boyte, Joshua Picotte, Daniel M. Howard, Kelcy Smith, and Kurtis J. Nelson. 2016. "An Optimal Sample Data Usage Strategy to Minimize Overfitting and Underfitting Effects in Regression Tree Models Based on Remotely-Sensed Data" Remote Sensing 8, no. 11: 943. https://doi.org/10.3390/rs8110943