Biomass Estimation Using 3D Data from Unmanned Aerial Vehicle Imagery in a Tropical Woodland
Abstract
:1. Introduction
2. Materials and Methods
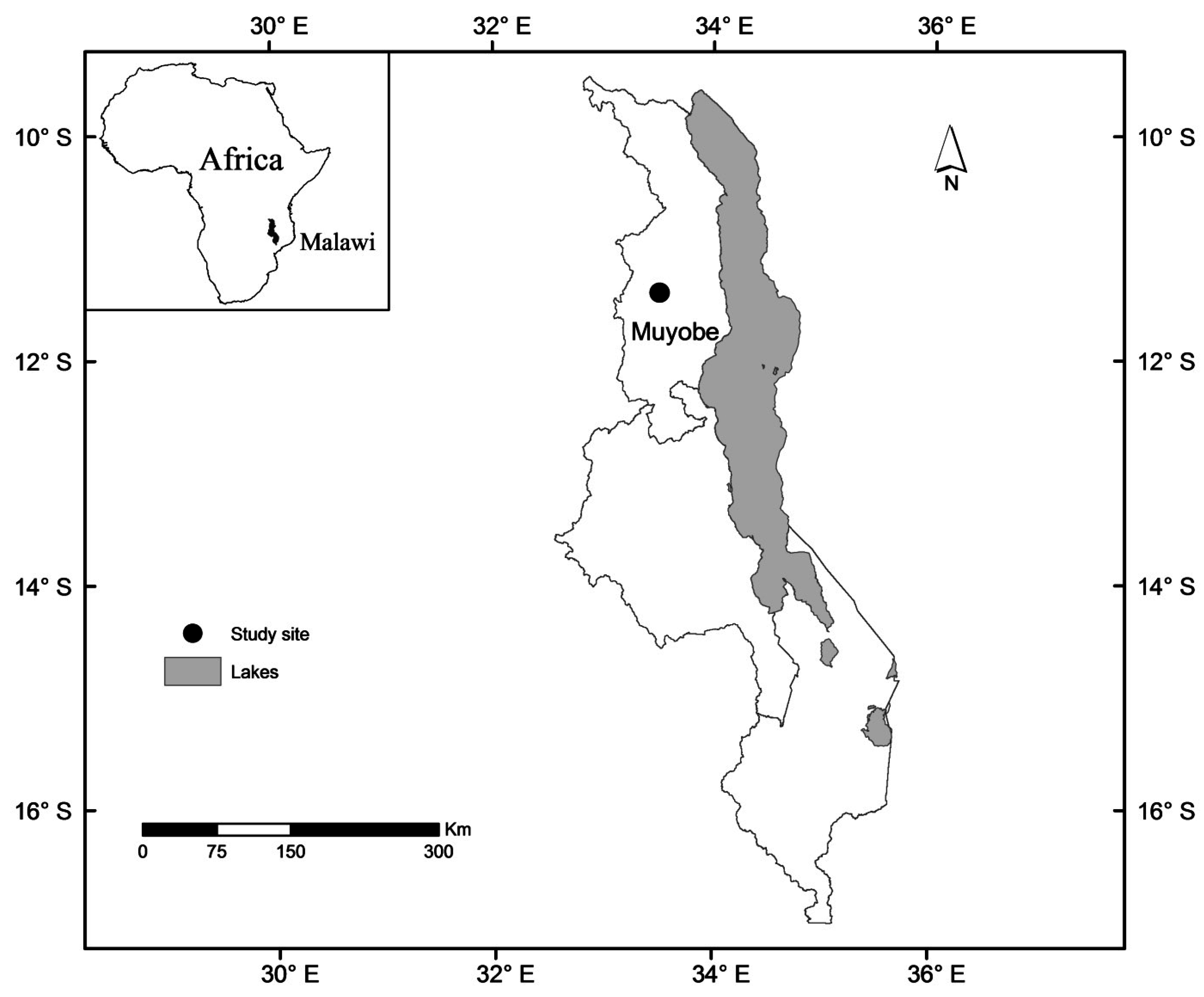
2.1. Study Area
2.2. Data Collection
2.2.1. Sampling Design and Ground Reference Data Collection
2.2.2. UAV Imagery Collection
2.3. Image Processing
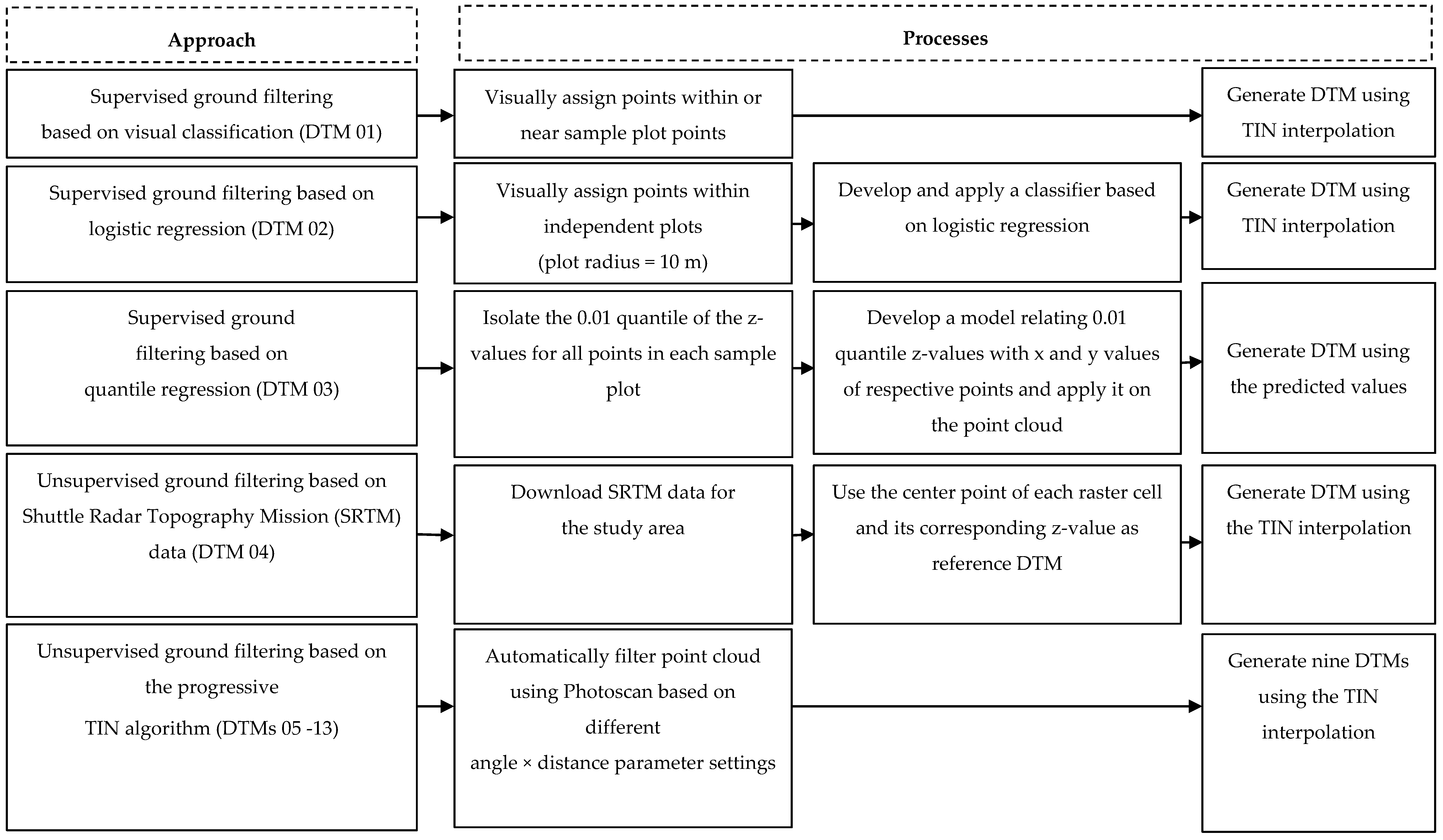
2.4. DTM Generation Methods
2.4.1. Supervised Ground Filtering Based on Visual Classification
2.4.2. Supervised Ground Filtering Based on Logistic Regression
2.4.3. Supervised Ground Filtering Based on Quantile Regression
2.4.4. Unsupervised Ground Filtering Based on Shuttle Radar Topography Mission (SRTM).
2.4.5. Unsupervised Ground Filtering Based on the Progressive TIN Algorithm
2.5. Variable Extraction
2.6. Model Development and Evaluation
3. Results
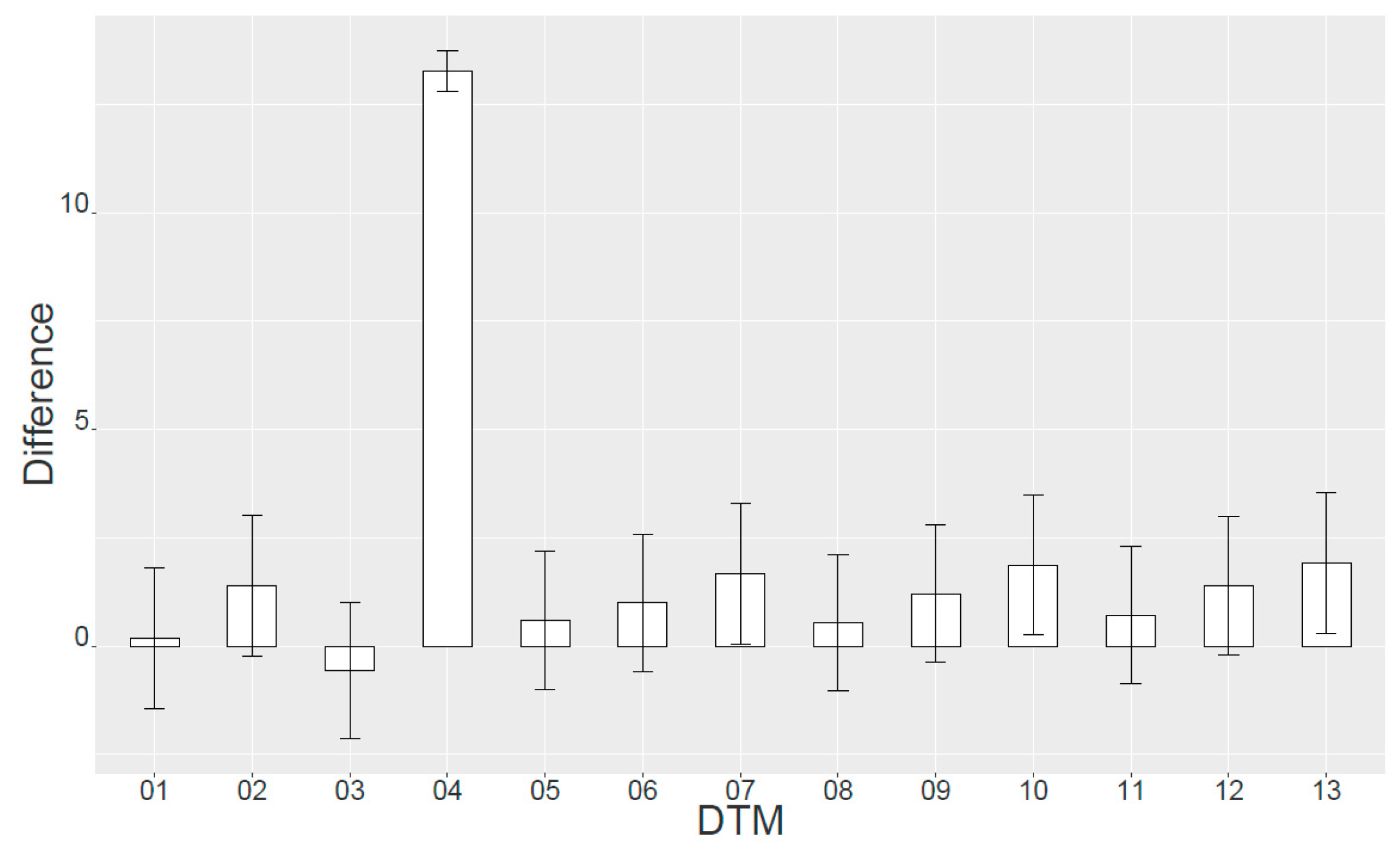
3.1. Comparison of the DTM Generation Methods
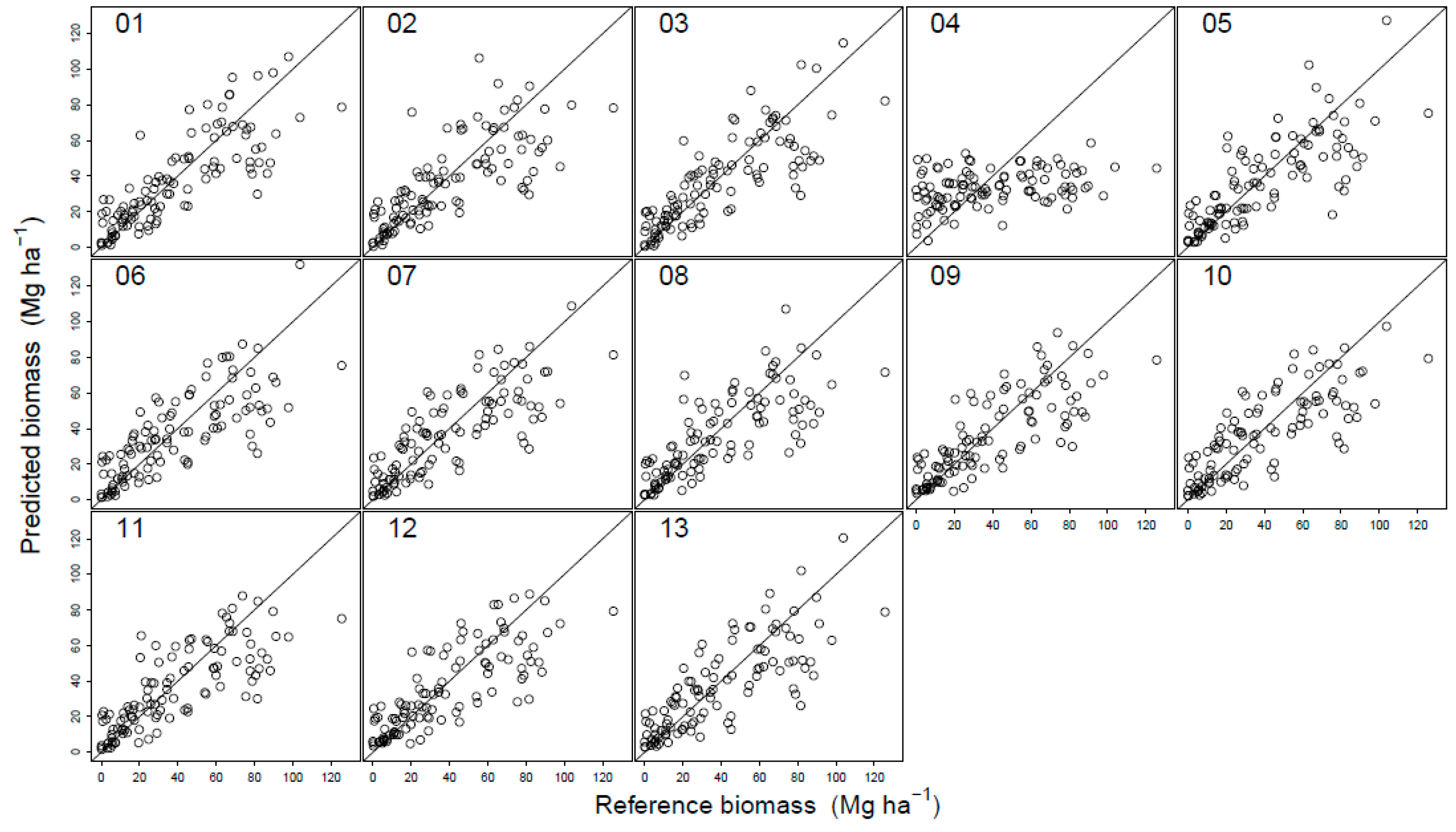
3.2. Regression Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gibbs, H.K.; Brown, S.; Niles, J.O.; Foley, J.A. Monitoring and estimating tropical forest carbon stocks: Making REDD a reality. Environ. Res. Lett. 2007, 2, 045023. [Google Scholar] [CrossRef]
- Government of Malawi. Government of Malawi REDD+ Action Plan 2014–2019; Department of Forestry, Ministry of Environment and Climate Change Management: Lilongwe, Malawi, 2015.
- Tomppo, E.; Malimbwi, R.; Katila, M.; Mäkisara, K.; Henttonen, H.M.; Chamuya, N.; Zahabu, E.; Otieno, J. A sampling design for a large area forest inventory: Case Tanzania. Can. J. For. Res. 2014, 44, 931–948. [Google Scholar] [CrossRef]
- Government of Malawi. Forest Resource Mapping Project under the Japanese Grant for the Forest Preservation Programme to the Republic of Malawi; Department of Forestry, Ministry of Environment and Climate Change Management: Lilongwe, Malawi, 2012.
- Chidumayo, E.N. Miombo Ecology and Management: An Introduction; IT Publications in Association with the Stockholm Environment Institute: London, UK, 1997; p. 166. [Google Scholar]
- Government of Malawi. Malawi State of Environment and Outlook Report; Ministry of Natural Resources, Energy and Environment: Lilongwe, Malawi, 2010.
- Angelsen, A. Moving Ahead with REDD: Issues, Options and Implications; Center for International Forestry Research (CIFOR): Bogor, Indonesia, 2008. [Google Scholar]
- DeFries, R.; Achard, F.; Brown, S.; Herold, M.; Murdiyarso, D.; Schlamadinger, B.; de Souza, C., Jr. Earth observations for estimating greenhouse gas emissions from deforestation in developing countries. Environ. Sci. Policy 2007, 10, 385–394. [Google Scholar] [CrossRef]
- Sinha, S.; Jeganathan, C.; Sharma, L.; Nathawat, M. A review of radar remote sensing for biomass estimation. Int. J. Environ. Sci. Technol. 2015, 12, 1779–1792. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Evaluating the utility of the medium-spatial resolution landsat 8 multispectral sensor in quantifying aboveground biomass in uMgeni catchment, South Africa. ISPRS J. Photogramm. Remote Sens. 2015, 101, 36–46. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T.; Bollandsås, O.M. Indirect and direct estimation of forest biomass change using forest inventory and airborne laser scanning data. Remote Sens. Environ. 2015, 164, 36–42. [Google Scholar] [CrossRef]
- Masek, J.G.; Hayes, D.J.; Joseph, H.M.; Healey, S.P.; Turner, D.P. The role of remote sensing in process-scaling studies of managed forest ecosystems. For. Ecol. Manag. 2015, 355, 109–123. [Google Scholar] [CrossRef]
- Barrett, F.; McRoberts, R.E.; Tomppo, E.; Cienciala, E.; Waser, L.T. A questionnaire-based review of the operational use of remotely sensed data by national forest inventories. Remote Sens. Environ. 2016, 174, 279–289. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T.; Bollandsås, O.M.; Gregoire, T.G.; Nelson, R.; Ståhl, G. Comparison of precision of biomass estimates in regional field sample surveys and airborne lidar-assisted surveys in Hedmark county, Norway. Remote Sens. Environ. 2013, 130, 108–120. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 1–43. [Google Scholar] [CrossRef]
- Kumar, L.; Sinha, P.; Taylor, S.; Alqurashi, A.F. Review of the use of remote sensing for biomass estimation to support renewable energy generation. J. Appl. Remote Sens. 2015, 9, 29. [Google Scholar] [CrossRef]
- Dandois, J.; Olano, M.; Ellis, E. Optimal altitude, overlap, and weather conditions for computer vision UAV estimates of forest structure. Remote Sens. 2015, 7, 13895–13920. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Skowronski, N.S.; Clark, K.L.; Gallagher, M.; Birdsey, R.A.; Hom, J.L. Airborne laser scanner-assisted estimation of aboveground biomass change in a temperate oak-pine forest. Remote Sens. Environ. 2014, 151, 166–174. [Google Scholar] [CrossRef]
- Patenaude, G.; Hill, R.A.; Milne, R.; Gaveau, D.L.A.; Briggs, B.B.J.; Dawson, T.P. Quantifying forest above ground carbon content using lidar remote sensing. Remote Sens. Environ. 2004, 93, 368–380. [Google Scholar] [CrossRef]
- Gonzalez, P.; Asner, G.P.; Battles, J.J.; Lefsky, M.A.; Waring, K.M.; Palace, M. Forest carbon densities and uncertainties from lidar, quickbird, and field measurements in California. Remote Sens. Environ. 2010, 114, 1561–1575. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Clark, D.B.; Knox, R.G.; Blair, J.B.; Hofton, M.A.; Chazdon, R.L.; Weishampel, J.F.; Prince, S. Estimation of tropical forest structural characteristics using large-footprint lidar. Remote Sens. Environ. 2002, 79, 305–319. [Google Scholar] [CrossRef]
- Mauya, E.; Hansen, E.; Gobakken, T.; Bollandsas, O.; Malimbwi, R.; Naesset, E. Effects of field plot size on prediction accuracy of aboveground biomass in airborne laser scanning-assisted inventories in tropical rain forests of Tanzania. Carbon Balance Manag. 2015, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vauhkonen, J.; Maltamo, M.; McRoberts, R.E.; Næsset, E. Introduction to forestry applications of airborne laser scanning. In Forestry Applications of Airborne Laser Scanning-Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 1–6. [Google Scholar]
- Whitehead, K.; Hugenholtz, C.H. Remote sensing of the environment with small unmanned aircraft systems (UASs), part 1: A review of progress and challenges. J. Unmanned Veh. Syst. 2014, 2, 69–85. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. High spatial resolution three-dimensional mapping of vegetation spectral dynamics using computer vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef]
- Puliti, S.; Ørka, H.; Gobakken, T.; Næsset, E. Inventory of small forest areas using an unmanned aerial system. Remote Sens. 2015, 7, 9632–9654. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Shao, G. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Malenovský, Z.; Turner, D.; Vopěnka, P. Assessment of forest structure using two UAV techniques: A comparison of airborne laser scanning and structure from motion (SfM) point clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef]
- Siebert, S.; Teizer, J. Mobile 3D mapping for surveying earthwork projects using an unmanned aerial vehicle (UAV) system. Autom. Constr. 2014, 41, 1–14. [Google Scholar] [CrossRef]
- Getzin, S.; Wiegand, K.; Schöning, I. Assessing biodiversity in forests using very high-resolution images and unmanned aerial vehicles. Methods Ecol. Evol. 2012, 3, 397–404. [Google Scholar] [CrossRef]
- Chidumayo, E.N. Forest degradation and recovery in a miombo woodland landscape in Zambia: 22 years of observations on permanent sample plots. For. Ecol. Manag. 2013, 291, 154–161. [Google Scholar] [CrossRef]
- Chidumayo, E.N.; Gumbo, D.J. The Dry Forests and Woodlands of Africa: Managing for Products and Services; Earthscan: London, UK, 2010. [Google Scholar]
- Næsset, E.; Økland, T. Estimating tree height and tree crown properties using airborne scanning laser in a boreal nature reserve. Remote Sens. Environ. 2002, 79, 105–115. [Google Scholar] [CrossRef]
- Ravindranath, N.H.; Ostwald, M. Carbon Inventory Methods—Handbook for Greenhouse Gas Inventory, Carbon Mitigation and Roundwood Production Projects; Springer: Heidelberg, Germany, 2008; Volume 29. [Google Scholar]
- Mauya, E.; Ene, L.; Bollandsås, O.; Gobakken, T.; Næsset, E.; Malimbwi, R.; Zahabu, E. Modelling aboveground forest biomass using airborne laser scanner data in the miombo woodlands of Tanzania. Carbon Balance Manag. 2015, 10, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregoire, T.G.; Næsset, E.; McRoberts, R.E.; Ståhl, G.; Andersen, H.-E.; Gobakken, T.; Ene, L.; Nelson, R. Statistical rigor in lidar-assisted estimation of aboveground forest biomass. Remote Sens. Environ. 2016, 173, 98–108. [Google Scholar] [CrossRef]
- Yang, J.; Prince, S.D. Remote sensing of savanna vegetation changes in eastern Zambia 1972–1989. Int. J. Remote Sens. 2000, 21, 301–322. [Google Scholar] [CrossRef]
- Fuller, D.O.; Prince, S.D.; Astle, W.L. The influence of canopy strata on remotely sensed observations of savanna-woodlands. Int. J. Remote Sens. 1997, 18, 2985–3009. [Google Scholar] [CrossRef]
- Maguya, A.; Junttila, V.; Kauranne, T. Algorithm for extracting digital terrain models under forest canopy from airborne lidar data. Remote Sens. 2014, 6, 6524–6548. [Google Scholar] [CrossRef]
- Niethammer, U.; James, M.R.; Rothmund, S.; Travelletti, J.; Joswig, M. UAV-based remote sensing of the super-sauze landslide: Evaluation and results. Eng. Geol. 2012, 128, 2–11. [Google Scholar] [CrossRef]
- Soininen, A. Terrascan User’s Guide; Terrasolid: Helsinki, Finland, 2004. [Google Scholar]
- McGaughey, R. FUSION/LDV: Software for LIDAR Data Analysis and Visualization; version 3.50; United States Department of Agriculture (USDA), Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 2015.
- Hardcastle, P.D. A Preliminary Silvicultural Classification of Malawi; Forestry Research Institute of Malawi: Zomba, Malawi, 1978. [Google Scholar]
- Kouba, J. A Guide to Using International Gnss Service (IGS) Products; Geodetic Survey Division, Natural Resources Canada: Ottawa, ON, Canada, 2015. [Google Scholar]
- Takasu, T. RTKLIB: Open Source Program Package for RTK-GPS; Free and Open Source Software for Geospatial Sydney; Open Source Geospatial Foundation (OSGeo): Sydney, Australia, 2009. [Google Scholar]
- Kachamba, D.J.; Eid, T.; Gobakken, T. Above- and belowground biomass models for trees in the miombo woodlands of Malawi. Forests 2016, 7, 38. [Google Scholar] [CrossRef]
- Sensefly. Ebee Sensefly: Extended User Manual; Sensefly, Ltd.: Cheseaux-Lausanne, Switzerland, 2015. [Google Scholar]
- AgiSoft. Agisoft Photoscan User Manual: Professional Edition, version 1.1; AgiSoft LLC: St. Petersburg, Russia, 2015. [Google Scholar]
- Stal, C.; Briese, C.; De Maeyer, P.; Dorninger, P.; Nuttens, T.; Pfeifer, N.; De Wulf, A. Classification of airborne laser scanning point clouds based on binomial logistic regression analysis. Int. J. Remote Sens. 2014, 35, 3219–3236. [Google Scholar] [CrossRef] [Green Version]
- Cade, B.S.; Noon, B.R. A gentle introduction to quantile regression for ecologists. Front. Ecol. Environ. 2003, 1, 412–420. [Google Scholar] [CrossRef]
- Fahrmeir, L.; Kneib, T.; Lang, S.; Marx, B. Regression; Springer: Berlin/Heidelberg, Germany, 2013; p. 698. [Google Scholar]
- Van Zyl, J.J. The shuttle radar topography mission (srtm): A breakthrough in remote sensing of topography. Acta Astronaut. 2001, 48, 559–565. [Google Scholar] [CrossRef]
- Axelsson, P. Processing of laser scanner data-algorithms and applications. ISPRS J. Photogramm. Remote Sens. 1999, 54, 138–147. [Google Scholar] [CrossRef]
- Næsset, E. Practical large-scale forest stand inventory using a small-footprint airborne scanning laser. Scand. J. For. Res. 2004, 19, 164–179. [Google Scholar] [CrossRef]
- Næsset, E.; Bollandsås, O.M.; Gobakken, T. Comparing regression methods in estimation of biophysical properties of forest stands from two different inventories using laser scanner data. Remote Sens. Environ. 2005, 94, 541–553. [Google Scholar] [CrossRef]
- White, J.; Wulder, M.; Varhola, A.; Vastaranta, M.; Coops, N.; Cook, B.; Pitt, D.; Woods, M. A Best Practices Guide for Generating Forest Inventory Attributes from Airborne Laser Scanning Data Using an Area-Based Approach (Version 2.0); Natural Resources Canada: Ottawa, ON, Canada, 2013; p. 50. [Google Scholar]
- Lumley, T. Leaps: Regression subset selection. In R Package, version 2.9; R Core Team: Vienna, Austria, 2009. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Meng, J.; Li, S.; Wang, W.; Liu, Q.; Xie, S.; Ma, W. Mapping forest health using spectral and textural information extracted from spot-5 satellite images. Remote Sens. 2016, 8, 719. [Google Scholar] [CrossRef]
- Snowdon, P. A ratio estimator for bias correction in logarithmic regressions. Can. J. For. Res. 1991, 21, 720–724. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning with Application in R; Springer: New York, NY, USA, 2013; Volume 103. [Google Scholar]
- Zhang, J.; Hu, J.; Lian, J.; Fan, Z.; Ouyang, X.; Ye, W. Seeing the forest from drones: Testing the potential of lightweight drones as a tool for long-term forest monitoring. Biol. Conserv. 2016, 198, 60–69. [Google Scholar] [CrossRef]
- Chianucci, F.; Disperati, L.; Guzzi, D.; Bianchini, D.; Nardino, V.; Lastri, C.; Rindinella, A.; Corona, P. Estimation of canopy attributes in beech forests using true colour digital images from a small fixed-wing UAV. Int. J. Appl. Earth Obs. Geoinf. 2016, 47, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, E.; Morris, C.; Belz, J. A global assessment of the srtm performance. Photogramm. Eng. Remote Sens. 2006, 72, 249–260. [Google Scholar] [CrossRef]
- Karwel, A.; Ewiak, I. Silk road for information from imagery. In Estimation of the Accuracy of the SRTM Terrain Model on the Area of Poland; Chen, J., Jiang, J., Genderen, J.V., Eds.; International Society for Photogrammetry and Remote Sensing: Beijing, China, 2008; pp. 169–172. [Google Scholar]
- Hofton, M.; Dubayah, R.; Blair, J.; Rabine, D. Validation of srtm elevations over vegetated and non-vegetated terrain using medium footprint lidar. Photogramm. Eng. Remote Sens. 2006, 72, 279–285. [Google Scholar] [CrossRef]
- Baltsavias, E.P. A comparison between photogrammetry and laser scanning. ISPRS J. Photogramm. Remote Sens. 1999, 54, 83–94. [Google Scholar] [CrossRef]
- Gobakken, T.; Bollandsås, O.M.; Næsset, E. Comparing biophysical forest characteristics estimated from photogrammetric matching of aerial images and airborne laser scanning data. Scand. J. For. Res. 2015, 30, 73–86. [Google Scholar] [CrossRef]
- Campbell, J.B.; Wynne, R.H. Introduction to Remote Sensing; Guilford Press: New York, NY, USA, 2011. [Google Scholar]
- Immitzer, M.; Stepper, C.; Böck, S.; Straub, C.; Atzberger, C. Use of worldview-2 stereo imagery and national forest inventory data for wall-to-wall mapping of growing stock. For. Ecol. Manag. 2016, 359, 232–246. [Google Scholar] [CrossRef]
- Frazer, G.; Magnussen, S.; Wulder, M.; Niemann, K. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of lidar-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Keller, M.; Palace, M.; Hurtt, G. Biomass estimation in the tapajos national forest, brazil: Examination of sampling and allometric uncertainties. For. Ecol. Manag. 2001, 154, 371–382. [Google Scholar] [CrossRef]
- Hansen, E.; Gobakken, T.; Bollandsas, O.; Zahabu, E.; Naesset, E. Modeling aboveground biomass in dense tropical submontane rainforest using airborne laser scanner data. Remote Sens. 2015, 7, 788–807. [Google Scholar] [CrossRef] [Green Version]
- Zahawi, R.A.; Dandois, J.P.; Holl, K.D.; Nadwodny, D.; Reid, J.L.; Ellis, E.C. Using lightweight unmanned aerial vehicles to monitor tropical forest recovery. Biol. Conserv. 2015, 186, 287–295. [Google Scholar] [CrossRef]

| Characteristic | Ground Reference Values | ||||
|---|---|---|---|---|---|
| Range | Mean | Std 1 | Cv 2 | Stderr 3 | |
| Biomass (Mg·ha−1) | 0–125.59 | 38.99 | 29.49 | 75.62 | 2.85 |
| Basal area (m2·ha−1) | 0–16.10 | 5.32 | 3.78 | 71.06 | 0.37 |
| Number of stems (ha−1) | 0–830 | 337 | 178 | 53 | 17 |
| Lorey’s mean height (m) | 3.76–14.58 | 8.81 | 2.41 | 27.31 | 0.23 |
| Date | Number of Flights | Number of Images | Flight Time (min) | Wind Speed (m·s−1) | Cloud Cover (%) |
|---|---|---|---|---|---|
| 23 April 2015 | 8 | 1241 | 153 | 6.0–9.5 | 10–80 |
| 24 April 2015 | 7 | 1301 | 146 | 6.0–9.0 | 20–80 |
| 25 April 2015 | 6 | 1118 | 132 | 5.0–9.0 | 10–100 |
| 26 April 2015 | 1 | 273 | 26 | 3.0–4.0 | 50 |
| Task | Parameters |
|---|---|
| (a) Image alignment | Accuracy: high |
| Pair selection: reference | |
| Key points: 40,000 | |
| Tie points: 1000 | |
| (b) Mesh building | Surface type: height field |
| Source data: dense cloud | |
| Face count: high | |
| (c) Guided marker positioning | Manual positioning of markers on the 14 GCPs for all the photos where a GCP was visible |
| (d) Building dense point cloud | Quality: medium |
| Depth filtering: mild |
| DTM 1 | Independent Variables 2 | r2 | Predicted Biomass Mg·ha−1 | RMSE | MPE | p-Value | ||
|---|---|---|---|---|---|---|---|---|
| Mg·ha−1 | % | Mg·ha−1 | % | |||||
| 01 | Hmax, D9, Ssd.blue | 0.67 | 39.64 | 18.36 | 46.8 | −0.41 | −1.1 | 0.82 |
| 02 | Hmax, D2, Ssd.green | 0.58 | 39.26 | 21.44 | 55.0 | −0.27 | −0.7 | 0.90 |
| 03 | Hmax, D5, Ssd.blue | 0.65 | 39.21 | 18.73 | 48.0 | −0.22 | −0.6 | 0.91 |
| 04 | S50.red | 0.12 | 38.95 | 31.87 | 81.7 | 0.04 | 0.1 | 0.99 |
| 05 | Hsd, D0, Ssd.blue | 0.61 | 39.44 | 20.38 | 52.2 | −0.45 | −1.2 | 0.82 |
| 06 | Hmax, S70.red, S90.green | 0.61 | 39.15 | 19.76 | 50.7 | −0.16 | −0.4 | 0.93 |
| 07 | Hmax, S70.red, S90.green | 0.64 | 39.03 | 18.21 | 46.7 | −0.03 | −0.1 | 0.99 |
| 08 | Hmax, D0, Ssd.blue | 0.59 | 38.60 | 22.73 | 58.3 | 0.39 | 1.0 | 0.86 |
| 09 | Hmax, D0, Scv.red | 0.63 | 38.90 | 20.40 | 52.3 | 0.09 | 0.2 | 0.96 |
| 10 | Hmax, S70.red, S90.green | 0.62 | 39.02 | 19.54 | 50.1 | −0.03 | −0.1 | 0.99 |
| 11 | Hmax, D0, Scv.green | 0.62 | 39.03 | 20.36 | 52.2 | −0.03 | −0.1 | 0.99 |
| 12 | Hmax, D0, Scv.red | 0.62 | 38.90 | 19.68 | 50.5 | 0.09 | 0.2 | 0.96 |
| 13 | Hmax, D0, S70.red, S90.green | 0.63 | 39.69 | 20.19 | 51.8 | −0.71 | −1.8 | 0.72 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kachamba, D.J.; Ørka, H.O.; Gobakken, T.; Eid, T.; Mwase, W. Biomass Estimation Using 3D Data from Unmanned Aerial Vehicle Imagery in a Tropical Woodland. Remote Sens. 2016, 8, 968. https://doi.org/10.3390/rs8110968
Kachamba DJ, Ørka HO, Gobakken T, Eid T, Mwase W. Biomass Estimation Using 3D Data from Unmanned Aerial Vehicle Imagery in a Tropical Woodland. Remote Sensing. 2016; 8(11):968. https://doi.org/10.3390/rs8110968
Chicago/Turabian StyleKachamba, Daud Jones, Hans Ole Ørka, Terje Gobakken, Tron Eid, and Weston Mwase. 2016. "Biomass Estimation Using 3D Data from Unmanned Aerial Vehicle Imagery in a Tropical Woodland" Remote Sensing 8, no. 11: 968. https://doi.org/10.3390/rs8110968







