A Review: Epigenetic Mechanism in Ochratoxin A Toxicity Studies
Abstract
:1. Introduction of Mycotoxins and Ochratoxin A
1.1. Mycotoxins
1.2. Ochratoxin A
2. Main Mechanisms of Ochratoxin A–Induced Toxicity
2.1. Oxidative Stress
2.2. Cell Apoptosis
2.3. Cell Autophagy
2.4. Calcium Homeostasis
2.5. DNA Adduct
2.6. Protein Synthesis Inhibition
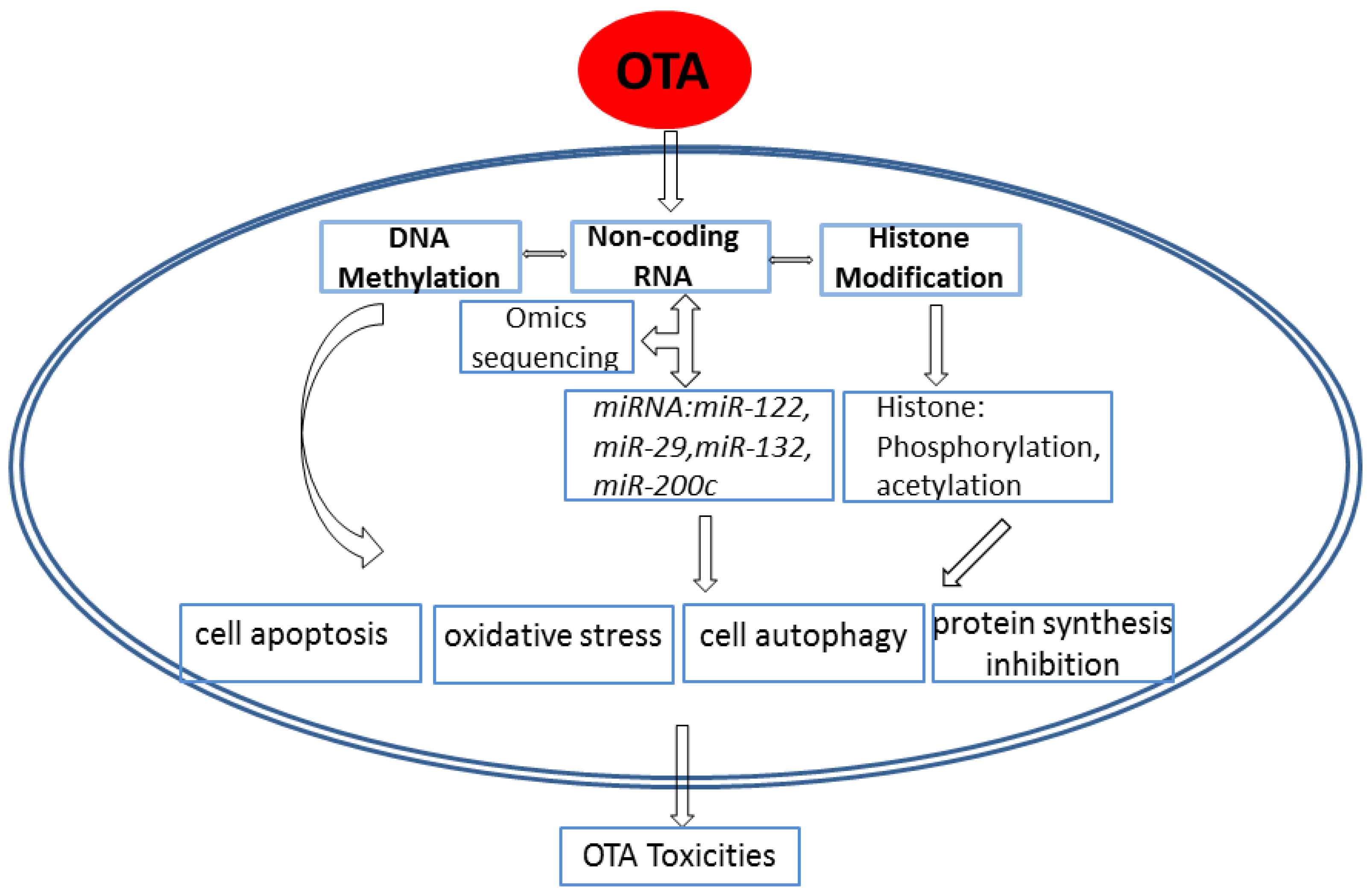
3. Advances in the Epigenetic Mechanism of Ochratoxin A–Induced Toxicity
3.1. Effects of OTA on DNA Methylation
3.2. Effects of OTA on Non-Coding RNA
3.3. Effects of OTA on Histone Modification
4. Conclusions and Future Perspectives
- (1)
- The specific target site of OTA should be detected by advanced technologies such as single cell sequencing, smart sequencing, reduced representation bisulfite sequencing (RRBS), and small RNA sequencing. These will provide more significant data for the understanding of OTA MOA.
- (1)
- ceRNA can be explored with OTA. Recently, the ceRNA hypothesis was proposed. mRNA and lncRNA or circRNA may bind competitively to miRNA to regulate the expression. This may provide a new field for us to understand the OTA MOA.
Acknowledgments
Conflicts of Interest
References
- Vettorazzi, A.; van Delft, J.; Lopez de Cerain, A. A review on Ochratoxin A transcriptomic studies. Food Chem. Toxicol. 2013, 59, 766–783. [Google Scholar] [CrossRef] [PubMed]
- Sorrenti, V.; Di Giacomo, C.; Acquaviva, R.; Barbagallo, I.; Bognanno, M.; Galvano, F. Toxicity of Ochratoxin A and its modulation by antioxidants: A review. Toxins 2013, 5, 1742–1766. [Google Scholar] [CrossRef] [PubMed]
- Herrman, J.; Walker, R. Risk analysis of mycotoxins by the joint FAO/WHO expert committee on food additives (JECFA). Food Nutr. Agric. 1999, 17–24. [Google Scholar]
- Van der Merwe, K.J.; Steyn, P.S.; Fourie, L.; Scott, D.B.; Theron, J.J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus wilh. Nature 1965, 205, 1112–1113. [Google Scholar] [CrossRef] [PubMed]
- Van Walbeek, W.; Scott, P.M.; Harwig, J.; Lawrence, J.W. Penicillium viridicatum westling: A new source of Ochratoxin A. Can. J. Microbiol. 1969, 15, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef] [PubMed]
- Bui-Klimke, T.R.; Wu, F. Ochratoxin A and human health risk: A review of the evidence. Crit. Rev. Food Sci. Nutr. 2015, 55, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Kamp, H.G.; Eisenbrand, G.; Schlatter, J.; Wurth, K.; Janzowski, C. Ochratoxin A: Induction of (oxidative) DNA damage, cytotoxicity and apoptosis in mammalian cell lines and primary cells. Toxicology 2005, 206, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Cavin, C.; Delatour, T.; Marin-Kuan, M.; Fenaille, F.; Holzhauser, D.; Guignard, G.; Bezencon, C.; Piguet, D.; Parisod, V.; Richoz-Payot, J.; et al. Ochratoxin A-mediated DNA and protein damage: Roles of nitrosative and oxidative stresses. Toxicol. Sci. 2009, 110, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Marin-Kuan, M.; Ehrlich, V.; Delatour, T.; Cavin, C.; Schilter, B. Evidence for a role of oxidative stress in the carcinogenicity of Ochratoxin A. J. Toxicol. 2011, 2011, 645361. [Google Scholar] [CrossRef] [PubMed]
- Petrik, J.; Zanic-Grubisic, T.; Barisic, K.; Pepeljnjak, S.; Radic, B.; Ferencic, Z.; Cepelak, I. Apoptosis and oxidative stress induced by Ochratoxin A in rat kidney. Arch. Toxicol. 2003, 77, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, G.J.; Nijmeijer, S.M.; Maas, R.F.; Roestenberg, P.; de Groene, E.M.; Fink-Gremmels, J. The role of oxidative stress in the Ochratoxin A-mediated toxicity in proximal tubular cells. Biochim. Biophys. Acta 2002, 1588, 149–158. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Y.; Xu, W.; Luo, Y.; Hao, J.; Shen, X.L.; Yang, X.; Li, X.; Huang, K. Zinc protects HepG2 cells against the oxidative damage and DNA damage induced by Ochratoxin A. Toxicol. Appl. Pharm. 2013, 268, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.G.; Saraiva, N.; Guerreiro, P.S.; Louro, H.; Silva, M.J.; Miranda, J.P.; Castro, M.; Batinic-Haberle, I.; Fernandes, A.S.; Oliveira, N.G. Ochratoxin A-induced cytotoxicity, genotoxicity and reactive oxygen species in kidney cells: An integrative approach of complementary endpoints. Food Chem. Toxicol. 2016, 87, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Meki, A.R.; Hussein, A.A. Melatonin reduces oxidative stress induced by Ochratoxin A in rat liver and kidney. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2001, 130, 305–313. [Google Scholar] [CrossRef]
- Limonciel, A.; Jennings, P. A review of the evidence that Ochratoxin A is an Nrf2 inhibitor: Implications for nephrotoxicity and renal carcinogenicity. Toxins 2014, 6, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Stachurska, A.; Ciesla, M.; Kozakowska, M.; Wolffram, S.; Boesch-Saadatmandi, C.; Rimbach, G.; Jozkowicz, A.; Dulak, J.; Loboda, A. Cross-talk between microRNAs, nuclear factor E2-related factor 2, and heme oxygenase-1 in Ochratoxin A-induced toxic effects in renal proximal tubular epithelial cells. Mol. Nutr. Food Res. 2013, 57, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Atroshi, F.; Biese, I.; Saloniemi, H.; Ali-Vehmas, T.; Saari, S.; Rizzo, A.; Veijalainen, P. Significance of apoptosis and its relationship to antioxidants after Ochratoxin A administration in mice. J. Pharm. Pharm. Sci. 2000, 3, 281–291. [Google Scholar] [PubMed]
- Seegers, J.C.; Bohmer, L.H.; Kruger, M.C.; Lottering, M.L.; de Kock, M. A comparative study of Ochratoxin A-induced apoptosis in hamster kidney and HeLa cells. Toxicol. Appl. Pharmacol. 1994, 129, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, C.; Sharaf El Dein, O.; El Golli, E.; Abid-Essefi, S.; Brenner, C.; Lemaire, C.; Bacha, H. Different apoptotic pathways induced by zearalenone, T-2 toxin and Ochratoxin A in human hepatoma cells. Toxicology 2008, 254, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Klaric, M.S.; Rumora, L.; Ljubanovic, D.; Pepeljnjak, S. Cytotoxicity and apoptosis induced by fumonisin B1, beauvericin and Ochratoxin A in porcine kidney PK15 cells: Effects of individual and combined treatment. Arch. Toxicol. 2008, 82, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Gekle, M.; Schwerdt, G.; Freudinger, R.; Mildenberger, S.; Wilflingseder, D.; Pollack, V.; Dander, M.; Schramek, H. Ochratoxin A induces JNK activation and apoptosis in MDCK-C7 cells at nanomolar concentrations. J. Pharmacol. Exp. Ther. 2000, 293, 837–844. [Google Scholar] [PubMed]
- Schramek, H.; Wilflingseder, D.; Pollack, V.; Freudinger, R.; Mildenberger, S.; Gekle, M. Ochratoxin A-induced stimulation of extracellular signal-regulated kinases 1/2 is associated with madin-darby canine kidney-C7 cell dedifferentiation. J. Pharmacol. Exp. Ther. 1997, 283, 1460–1468. [Google Scholar] [PubMed]
- Ozcan, Z.; Gul, G.; Yaman, I. Ochratoxin A activates opposing c-MET/PI3K/AKt and MAPK/ERK 1–2 pathways in human proximal tubule HK-2 cells. Arch. Toxicol. 2015, 89, 1313–1327. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Shen, X.L.; Zhang, B.; Li, Y.; Xu, W.; Zhao, C.; Luo, Y.; Huang, K. Apoptosis signal-regulating kinase 1 promotes Ochratoxin A-induced renal cytotoxicity. Sci. Rep. 2015, 5, 8078. [Google Scholar] [CrossRef] [PubMed]
- Aleo, M.D.; Wyatt, R.D.; Schnellmann, R.G. Mitochondrial dysfunction is an early event in Ochratoxin A but not oosporein toxicity to rat renal proximal tubules. Toxicol. Appl. Pharmacol. 1991, 107, 73–80. [Google Scholar] [CrossRef]
- Shen, X.L.; Zhang, B.; Liang, R.; Cheng, W.H.; Xu, W.; Luo, Y.; Zhao, C.; Huang, K. Central role of Nix in the autophagic response to Ochratoxin A. Food Chem. Toxicol. 2014, 69, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Novak, I. Mitophagy: A complex mechanism of mitochondrial removal. Antioxid. Redox Signal. 2012, 17, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Rahimtula, A.D.; Chong, X. Alterations in calcium homeostasis as a possible cause of Ochratoxin A nephrotoxicity. IARC Sci. Publ. 1991, 207–214. [Google Scholar]
- Khan, S.; Martin, M.; Bartsch, H.; Rahimtula, A.D. Perturbation of liver microsomal calcium homeostasis by Ochratoxin A. Biochem. Pharmacol. 1989, 38, 67–72. [Google Scholar] [CrossRef]
- Dopp, E.; Muller, J.; Hahnel, C.; Schiffmann, D. Induction of genotoxic effects and modulation of the intracellular calcium level in syrian hamster embryo (SHE) fibroblasts caused by Ochratoxin A. Food Chem. Toxicol. 1999, 37, 713–721. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Chakor, K.; Creppy, E.E.; Dirheimer, G. DNA adduct formation in mice treated with Ochratoxin A. IARC Sci. Publ. 1991, 115, 245–253. [Google Scholar]
- Grosse, Y.; Baudrimont, I.; Castegnaro, M.; Betbeder, A.M.; Creppy, E.E.; Dirheimer, G.; Pfohl-Leszkowicz, A. Formation of Ochratoxin A metabolites and DNA-adducts in monkey kidney cells. Chem.-Biol. Interact. 1995, 95, 175–187. [Google Scholar] [CrossRef]
- El Adlouni, C.; Pinelli, E.; Azemar, B.; Zaoui, D.; Beaune, P.; Pfohl-Leszkowicz, A. Phenobarbital increases DNA adduct and metabolites formed by Ochratoxin A: Role of CYP 2C9 and microsomal glutathione-S-transferase. Environ. Mol. Mutagen. 2000, 35, 123–131. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Grosse, Y.; Kane, A.; Creppy, E.E.; Dirheimer, G. Differential DNA adduct formation and disappearance in three mouse tissues after treatment with the mycotoxin Ochratoxin A. Mutat. Res. 1993, 289, 265–273. [Google Scholar] [CrossRef]
- Miljkovic, A.; Pfohl-Leszkowicz, A.; Dobrota, M.; Mantle, P.G. Comparative responses to mode of oral administration and dose of Ochratoxin A or nephrotoxic extract of Penicillium polonicum in rats. Exp. Toxicol. Pathol. 2003, 54, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Petkova-Bocharova, T.; Stoichev, I.I.; Chernozemsky, I.N.; Castegnaro, M.; Pfohl-Leszkowicz, A. Formation of DNA adducts in tissues of mouse progeny through transplacental contamination and/or lactation after administration of a single dose of Ochratoxin A to the pregnant mother. Environ. Mol. Mutagen. 1998, 32, 155–162. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Grosse, Y.; Castegnaro, M.; Nicolov, I.G.; Chernozemsky, I.N.; Bartsch, H.; Betbeder, A.M.; Creppy, E.E.; Dirheimer, G. Ochratoxin A-related DNA adducts in urinary tract tumours of Bulgarian subjects. IARC Sci. Publ. 1993, 124, 141–148. [Google Scholar]
- Manderville, R.A. A case for the genotoxicity of Ochratoxin A by bioactivation and covalent DNA adduction. Chem. Res. Toxicol. 2005, 18, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Bendele, A.M.; Carlton, W.W.; Krogh, P.; Lillehoj, E.B. Ochratoxin A carcinogenesis in the (C57bl/6J × C3H)F1 mouse. J. Natl. Cancer Inst. 1985, 75, 733–742. [Google Scholar] [PubMed]
- Mantle, P.G.; Faucet-Marquis, V.; Manderville, R.A.; Squillaci, B.; Pfohl-Leszkowicz, A. Structures of covalent adducts between DNA and Ochratoxin A: A new factor in debate about genotoxicity and human risk assessment. Chem. Res. Toxicol. 2010, 23, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Mally, A.; Zepnik, H.; Wanek, P.; Eder, E.; Dingley, K.; Ihmels, H.; Volkel, W.; Dekant, W. Ochratoxin A: Lack of formation of covalent DNA adducts. Chem. Res. Toxicol. 2004, 17, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Mally, A.; Pepe, G.; Ravoori, S.; Fiore, M.; Gupta, R.C.; Dekant, W.; Mosesso, P. Ochratoxin A causes DNA damage and cytogenetic effects but no DNA adducts in rats. Chem. Res. Toxicol. 2005, 18, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Creppy, E.E.; Mayer, M.; Kern, D.; Schlegel, M.; Steyn, P.S.; Vleggaar, R.; Dirheimer, G. In vitro inhibition of yeast valyl-tRNA synthetase by the valine homologue of Ochratoxin A. Biochim. Biophys. Acta 1981, 656, 265–268. [Google Scholar] [CrossRef]
- Creppy, E.E.; Kern, D.; Steyn, P.S.; Vleggaar, R.; Roschenthaler, R.; Dirheimer, G. Comparative study of the effect of Ochratoxin A analogues on yeast aminoacyl-tRNA synthetases and on the growth and protein synthesis of hepatoma cells. Toxicol. Lett. 1983, 19, 217–224. [Google Scholar] [CrossRef]
- Creppy, E.E.; Stormer, F.C.; Kern, D.; Roschenthaler, R.; Dirheimer, G. Effects of ochratoxin a metabolites on yeast phenylalanyl-tRNA synthetase and on the growth and in vivo protein synthesis of hepatoma cells. Chem.-Biol. Interact. 1983, 47, 239–247. [Google Scholar] [CrossRef]
- Creppy, E.E.; Roschenthaler, R.; Dirheimer, G. Inhibition of protein synthesis in mice by Ochratoxin A and its prevention by phenylalanine. Food Chem. Toxicol. 1984, 22, 883–886. [Google Scholar] [CrossRef]
- Capuano, F.; Mulleder, M.; Kok, R.; Blom, H.J.; Ralser, M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other Yeast species. Anal. Chem. 2014, 86, 3697–3702. [Google Scholar] [CrossRef] [PubMed]
- Wing, M.R.; Devaney, J.M.; Joffe, M.M.; Xie, D.; Feldman, H.I.; Dominic, E.A.; Guzman, N.J.; Ramezani, A.; Susztak, K.; Herman, J.G.; et al. DNA methylation profile associated with rapid decline in kidney function: Findings from the CRIC study. Nephrol. Dial. Transplant. 2014, 29, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Giromini, C.; Rebucci, R.; Fusi, E.; Rossi, L.; Saccone, F.; Baldi, A. Cytotoxicity, apoptosis, DNA damage and methylation in mammary and kidney epithelial cell lines exposed to Ochratoxin A. Cell Biol. Toxicol. 2016, 32, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, J.; Huang, K.; Qi, X.; Dai, Q.; Mei, X.; Xu, W. Dynamic changes of global DNA methylation and hypermethylation of cell adhesion-related genes in rat kidneys in response to Ochratoxin A. World Mycotoxin J. 2015, 8, 465–476. [Google Scholar] [CrossRef]
- Dai, Q.; Zhao, J.; Qi, X.; Xu, W.; He, X.; Guo, M.; Dweep, H.; Cheng, W.H.; Luo, Y.; Xia, K.; et al. Microrna profiling of rats with Ochratoxin A nephrotoxicity. BMC Genom. 2014, 15, 333. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yang, X.; Chen, S.; He, X.; Dweep, H.; Guo, M.; Cheng, W.-H.; Xu, W.; Luo, Y.; Gretz, N.; et al. Ochratoxin A induced early hepatotoxicity: New mechanistic insights from microRNA, mRNA and proteomic profiling studies. Sci. Rep. 2014, 4, 5163. [Google Scholar] [CrossRef]
- Zhao, J.; Qi, X.; Dai, Q.; He, X.; Dweep, H.; Guo, M.; Luo, Y.; Gretz, N.; Luo, H.; Huang, K.; et al. Toxicity study of Ochratoxin A using HEK293 and HepG2 cell lines based on microRNA profiling. Hum. Exp. Toxicol. 2016, 36, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Hennemeier, I.; Humpf, H.U.; Gekle, M.; Schwerdt, G. Role of microRNA-29b in the Ochratoxin A-induced enhanced collagen formation in human kidney cells. Toxicology 2014, 324, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Nassirpour, R.; Mehta, P.P.; Yin, M.J. miR-122 regulates tumorigenesis in hepatocellular carcinoma by targeting AKT3. PLoS ONE 2013, 8, e79655. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yu, T.; Qi, X.; Yang, B.; Shi, L.; Luo, H.; He, X.; Huang, K.; Xu, W. miR-122 plays an important role in Ochratoxin A-induced hepatocyte apoptosis in vitro and in vivo. Toxicol. Res. 2016, 5, 160–167. [Google Scholar] [CrossRef]
- Chen, R.; Deng, L.; Yu, X.; Wang, X.; Zhu, L.; Yu, T.; Zhang, Y.; Zhou, B.; Xu, W.; Chen, L.; et al. miR-122 partly mediates the Ochratoxin A-induced GC-2 cell apoptosis. Toxicol. In Vitro 2015, 30, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Reinherz, E.L.; Reche, P.A. Recognition and classification of histones using support vector machine. J. Comput. Biol. 2006, 13, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liu, J.; An, S.; Nishino, T.; Hishikawa, Y.; Koji, T. Immunohistochemical analysis of histone H3 modifications in germ cells during mouse spermatogenesis. Acta Histochem. Cytochem. 2011, 44, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Czakai, K.; Muller, K.; Mosesso, P.; Pepe, G.; Schulze, M.; Gohla, A.; Patnaik, D.; Dekant, W.; Higgins, J.M.; Mally, A. Perturbation of mitosis through inhibition of histone acetyltransferases: The key to Ochratoxin A toxicity and carcinogenicity? Toxicol. Sci. 2011, 122, 317–329. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Zhang, B.; Dai, Y.; Li, H.; Xu, W. A Review: Epigenetic Mechanism in Ochratoxin A Toxicity Studies. Toxins 2017, 9, 113. https://doi.org/10.3390/toxins9040113
Zhu L, Zhang B, Dai Y, Li H, Xu W. A Review: Epigenetic Mechanism in Ochratoxin A Toxicity Studies. Toxins. 2017; 9(4):113. https://doi.org/10.3390/toxins9040113
Chicago/Turabian StyleZhu, Liye, Boyang Zhang, Yaqi Dai, Hongyu Li, and Wentao Xu. 2017. "A Review: Epigenetic Mechanism in Ochratoxin A Toxicity Studies" Toxins 9, no. 4: 113. https://doi.org/10.3390/toxins9040113




