Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases
Abstract
1. Introduction
2. Results
2.1. Patient and Tumor Characteristics
2.2. Molecular and Pathological Profiles of Colorectal Cancer with Brain Metastases
2.3. Concordance of Molecular and Pathological Profiles between Brain Metastases and Their Paired Primary Tumors
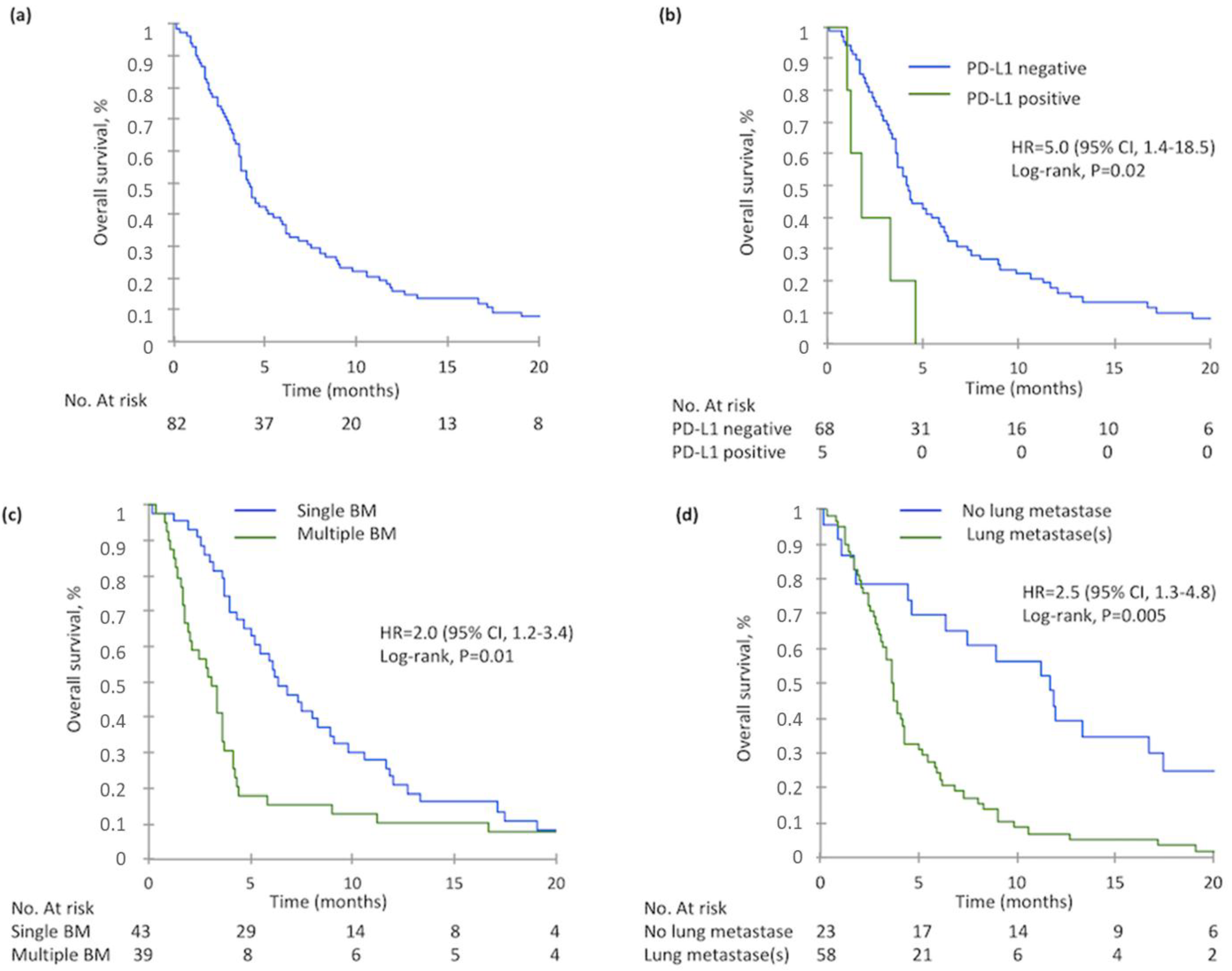
2.4. Overall Survival
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Molecular Analyses
4.3. Tissue Microarray Construction and Immunohistochemistry
4.4. Fluorescent In Situ Hybridization (FISH)
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | Brain Metastases |
| CI | Confidence Interval |
| CT-scan | Computed Tomography scan |
| CRC | Colorectal Cancer |
| ECM | Extracranial Metastasis(es) |
| ECOG | Eastern Cooperative Oncology Group score |
| FISH | Fluorescence In Situ Hybridization |
| HR | Hazard Ratio |
| IHC | Immunohistochemistry |
| OS | Overall Survival |
| mCRC | Metastatic Colorectal Cancer |
| MMR | Mismatch Repair |
| MRI | Magnetic Resonance Imaging |
| pMMR | Proficient Mismatch Repair |
| dMMR | Deficient Mismatch Repair |
| PPT | Paired Primary Tumor |
| TMA | Tissue Microarray |
| TILs | Tumor-Infiltrating Lymphocytes |
References
- Christensen, T.D.; Spindler, K.-L.G.; Palshof, J.A.; Nielsen, D.L. Systematic review: Brain metastases from colorectal cancer—Incidence and patient characteristics. BMC Cancer 2016, 16, 260. [Google Scholar] [CrossRef]
- Jung, M.; Ahn, J.B.; Chang, J.H.; Suh, C.O.; Hong, S.; Roh, J.K.; Shin, S.J.; Rha, S.Y. Brain metastases from colorectal carcinoma: Prognostic factors and outcome. J. Neurooncol. 2011, 101, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Tanriverdi, O.; Kaytan-Saglam, E.; Ulger, S.; Bayoglu, I.V.; Turker, I.; Ozturk-Topcu, T.; Cokmert, S.; Turhal, S.; Oktay, E.; Karabulut, B.; et al. The clinical and pathological features of 133 colorectal cancer patients with brain metastasis: A multicenter retrospective analysis of the Gastrointestinal Tumors Working Committee of the Turkish Oncology Group (TOG). Med. Oncol. 2014, 31, 152. [Google Scholar] [CrossRef] [PubMed]
- Yaeger, R.; Cowell, E.; Chou, J.F.; Gewirtz, A.N.; Borsu, L.; Vakiani, E.; Solit, D.B.; Rosen, N.; Capanu, M.; Ladanyi, M.; et al. RAS mutations affect pattern of metastatic spread and increase propensity for brain metastasis in colorectal cancer. Cancer 2015, 121, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.D.; Palshof, J.A.; Larsen, F.O.; Høgdall, E.; Poulsen, T.S.; Pfeiffer, P.; Jensen, B.V.; Yilmaz, M.K.; Christensen, I.J.; Nielsen, D. Risk factors for brain metastases in patients with metastatic colorectal cancer. Acta Oncol. 2017, 56, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Michl, M.; Thurmaier, J.; Schubert-Fritschle, G.; Wiedemann, M.; Laubender, R.P.; Nüssler, N.C.; Ruppert, R.; Kleeff, J.; Schepp, W.; Reuter, C.; et al. Brain Metastasis in Colorectal Cancer Patients: Survival and Analysis of Prognostic Factors. Clin. Colorectal. Cancer 2015, 14, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zeng, W.; Huang, C.; Wang, J.; Yang, D.; Ma, D. Predictive and Prognostic Implications of Mutation Profiling and Microsatellite Instability Status in Patients with Metastatic Colorectal Carcinoma. Available online: https://www.hindawi.com/journals/grp/2018/4585802/abs/ (accessed on 27 November 2018).
- Prasanna, T.; Karapetis, C.S.; Roder, D.; Tie, J.; Padbury, R.; Price, T.; Wong, R.; Shapiro, J.; Nott, L.; Lee, M.; et al. The survival outcome of patients with metastatic colorectal cancer based on the site of metastases and the impact of molecular markers and site of primary cancer on metastatic pattern. Acta Oncol. 2018, 57, 1438–1444. [Google Scholar] [CrossRef] [PubMed]
- Vakiani, E.; Janakiraman, M.; Shen, R.; Sinha, R.; Zeng, Z.; Shia, J.; Cercek, A.; Kemeny, N.; D’Angelica, M.; Viale, A.; et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J. Clin. Oncol. 2012, 30, 2956–2962. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Carter, S.L.; Santagata, S.; Cahill, D.P.; Taylor-Weiner, A.; Jones, R.T.; Van Allen, E.M.; Lawrence, M.S.; Horowitz, P.M.; Cibulskis, K.; et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015, 5, 1164–1177. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Oliner, K.S.; Siena, S.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013, 369, 1023–1034. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Schur, S.; Füreder, L.M.; Gatterbauer, B.; Dieckmann, K.; Widhalm, G.; Hainfellner, J.; Zielinski, C.C.; Birner, P.; Bartsch, R.; et al. Descriptive statistical analysis of a real life cohort of 2419 patients with brain metastases of solid cancers. ESMO Open 2016, 1, e000024. [Google Scholar] [CrossRef] [PubMed]
- El-Deiry, W.S.; Vijayvergia, N.; Xiu, J.; Scicchitano, A.; Lim, B.; Yee, N.S.; Harvey, H.A.; Gatalica, Z.; Reddy, S. Molecular profiling of 6,892 colorectal cancer samples suggests different possible treatment options specific to metastatic sites. Cancer Biol. Ther. 2015, 16, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Tougeron, D.; Fauquembergue, E.; Rouquette, A.; Le Pessot, F.; Sesboüé, R.; Laurent, M.; Berthet, P.; Mauillon, J.; Di Fiore, F.; Sabourin, J.-C.; et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod. Pathol. 2009, 22, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Maby, P.; Tougeron, D.; Hamieh, M.; Mlecnik, B.; Kora, H.; Bindea, G.; Angell, H.K.; Fredriksen, T.; Elie, N.; Fauquembergue, E.; et al. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015, 75, 3446–3455. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.H.; Cavalcanti, M.S.; Segal, N.H.; Hechtman, J.F.; Weiser, M.R.; Smith, J.J.; Garcia-Aguilar, J.; Sadot, E.; Ntiamoah, P.; Markowitz, A.J.; et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod. Pathol. 2016, 29, 1433–1442. [Google Scholar] [CrossRef]
- Zhang, M.; Li, G.; Sun, X.; Ni, S.; Tan, C.; Xu, M.; Huang, D.; Ren, F.; Li, D.; Wei, P.; et al. MET amplification, expression, and exon 14 mutations in colorectal adenocarcinoma. Hum. Pathol. 2018, 77, 108–115. [Google Scholar] [CrossRef]
- Abou-Bakr, A.A.; Elbasmi, A. c-MET overexpression as a prognostic biomarker in colorectal adenocarcinoma. Gulf. J. Oncol. 2013, 1, 28–34. [Google Scholar]
- Liu, Y.; Yu, X.-F.; Zou, J.; Luo, Z.-H. Prognostic value of c-Met in colorectal cancer: A meta-analysis. World J. Gastroenterol. 2015, 21, 3706–3710. [Google Scholar] [CrossRef]
- Zeng, Z.-S.; Weiser, M.R.; Kuntz, E.; Chen, C.-T.; Khan, S.A.; Forslund, A.; Nash, G.M.; Gimbel, M.; Yamaguchi, Y.; Culliford, A.T.; et al. c-Met gene amplification is associated with advanced stage colorectal cancer and liver metastases. Cancer Lett. 2008, 265, 258–269. [Google Scholar] [CrossRef]
- Aprile, G.; Casagrande, M.; De Maglio, G.; Fontanella, C.; Rihawi, K.; Bonotto, M.; Pisa, F.E.; Tuniz, F.; Pizzolitto, S.; Fasola, G. Comparison of the molecular profile of brain metastases from colorectal cancer and corresponding primary tumors. Future Oncol. 2017, 13, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Jeantet, M.; Tougeron, D.; Tachon, G.; Cortes, U.; Archambaut, C.; Fromont, G.; Karayan-Tapon, L. High Intra- and Inter-Tumoral Heterogeneity of RAS Mutations in Colorectal Cancer. Int. J. Mol. Sci. 2016, 17, 2015. [Google Scholar] [CrossRef] [PubMed]
- Feilchenfeldt, J.; Varga, Z.; Siano, M.; Grabsch, H.I.; Held, U.; Schuknecht, B.; Trip, A.; Hamaguchi, T.; Gut, P.; Balague, O.; et al. Brain metastases in gastro-oesophageal adenocarcinoma: Insights into the role of the human epidermal growth factor receptor 2 (HER2). Br. J. Cancer 2015, 113, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Gabos, Z.; Sinha, R.; Hanson, J.; Chauhan, N.; Hugh, J.; Mackey, J.R.; Abdulkarim, B. Prognostic significance of human epidermal growth factor receptor positivity for the development of brain metastasis after newly diagnosed breast cancer. J. Clin. Oncol. 2006, 24, 5658–5663. [Google Scholar] [CrossRef]
- Harter, P.N.; Bernatz, S.; Scholz, A.; Zeiner, P.S.; Zinke, J.; Kiyose, M.; Blasel, S.; Beschorner, R.; Senft, C.; Bender, B.; et al. Distribution and prognostic relevance of tumor-infiltrating lymphocytes (TILs) and PD-1/PD-L1 immune checkpoints in human brain metastases. Oncotarget 2015, 6, 40836–40849. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, A.S.; Aubry, M.C.; Moser, J.C.; Harrington, S.M.; Dronca, R.S.; Park, S.S.; Dong, H. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Ann. Oncol. 2016, 27, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Puhalla, S.; Elmquist, W.; Freyer, D.; Kleinberg, L.; Adkins, C.; Lockman, P.; McGregor, J.; Muldoon, L.; Nesbit, G.; Peereboom, D.; et al. Unsanctifying the sanctuary: Challenges and opportunities with brain metastases. Neuro-Oncology 2015, 17, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.B.; Gettinger, S.N.; Mahajan, A.; Chiang, A.C.; Herbst, R.S.; Sznol, M.; Tsiouris, A.J.; Cohen, J.; Vortmeyer, A.; Jilaveanu, L.; et al. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: Early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 976–983. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Aprile, G.; Rimassa, L.; Franco, P.; Lonardi, S.; Cremolini, C.; Biondani, P.; Sbicego, E.L.; Pasqualetti, F.; Tomasello, G.; et al. A new nomogram for estimating survival in patients with brain metastases secondary to colorectal cancer. Radiother. Oncol. 2015, 117, 315–321. [Google Scholar] [CrossRef]
- Droeser, R.A.; Hirt, C.; Viehl, C.T.; Frey, D.M.; Nebiker, C.; Huber, X.; Zlobec, I.; Eppenberger-Castori, S.; Tzankov, A.; Rosso, R.; et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur. J. Cancer 2013, 49, 2233–2242. [Google Scholar] [CrossRef]
- Téglási, V.; Reiniger, L.; Fábián, K.; Pipek, O.; Csala, I.; Bagó, A.G.; Várallyai, P.; Vízkeleti, L.; Rojkó, L.; Tímár, J.; et al. Evaluating the significance of density, localization, and PD-1/PD-L1 immunopositivity of mononuclear cells in the clinical course of lung adenocarcinoma patients with brain metastasis. Neuro-Oncology 2017, 19, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Cortes, U.; Guilloteau, K.; Rouvreau, M.; Archaimbault, C.; Villalva, C.; Karayan-Tapon, L. Development of pyrosequencing methods for the rapid detection of RAS mutations in clinical samples. Exp. Mol. Pathol. 2015, 99, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.-S.; Squire, J.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Mol. Diagn. 2013, 15, 415–453. [Google Scholar] [CrossRef] [PubMed]
- Houang, M.; Toon, C.W.; Clarkson, A.; Sioson, L.; de Silva, K.; Watson, N.; Singh, N.R.; Chou, A.; Gill, A.J. ALK and ROS1 overexpression is very rare in colorectal adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Yatabe, Y. ALK FISH and IHC: You cannot have one without the other. J. Thorac. Oncol. 2015, 10, 548–550. [Google Scholar] [CrossRef]
- Bardelli, A.; Corso, S.; Bertotti, A.; Hobor, S.; Valtorta, E.; Siravegna, G.; Sartore-Bianchi, A.; Scala, E.; Cassingena, A.; Zecchin, D.; et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013, 3, 658–673. [Google Scholar] [CrossRef]
- Valtorta, E.; Martino, C.; Sartore-Bianchi, A.; Penaullt-Llorca, F.; Viale, G.; Risio, M.; Rugge, M.; Grigioni, W.; Bencardino, K.; Lonardi, S.; et al. Assessment of a HER2 scoring system for colorectal cancer: Results from a validation study. Mod. Pathol. 2015, 28, 1481–1491. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Zito Marino, F.; Rocco, G.; Morabito, A.; Mignogna, C.; Intartaglia, M.; Liguori, G.; Botti, G.; Franco, R. A new look at the ALK gene in cancer: Copy number gain and amplification. Expert Rev. Anticancer Ther. 2016, 16, 493–502. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Oddo, D.; Gloghini, A.; Valtorta, E.; Berenato, R.; Barault, L.; Caporale, M.; Busico, A.; Morano, F.; Gualeni, A.V.; et al. MET-Driven Resistance to Dual EGFR and BRAF Blockade May Be Overcome by Switching from EGFR to MET Inhibition in BRAF-Mutated Colorectal Cancer. Cancer Discov. 2016, 6, 963–971. [Google Scholar] [CrossRef]
| Characteristics | Patients (n = 82) |
|---|---|
| Age at primary tumor diagnostic, years | |
| Median (range) | 64 (35–85) |
| Gender, n (%) | |
| Male | 52 (63) |
| Female | 30 (37) |
| Site of primary tumor, n (%) | |
| Ascending colon | 19 (23) |
| Descending colon | 24 (29) |
| Rectum | 35 (42) |
| Bifocal tumor | 5 (6) |
| Tumor grade, n (%) | |
| Well or moderately differentiated | 61 (87) |
| Poorly differentiated | 9 (13) |
| Missing | 12 |
| Stage at initial CRC diagnostic, n (%) | |
| I | 4 (5) |
| II | 13 (16) |
| III | 26 (32) |
| IV | 39 (47) |
| Primary tumor resection, n (%) | |
| No | 11 (13) |
| Yes | 71 (87) |
| ECOG performance status at BM diagnosis, n (%) | |
| < 2 | 43 (54) |
| ≥ 2 | 36 (46) |
| Missing | 3 |
| Number of BM, n (%) | |
| Single | 43 (52) |
| Multiple | 39 (48) |
| Site of BM, n (%) | |
| Supratentorial | 46 (56) |
| Subtentorial | 18 (22) |
| Both | 18 (22) |
| Delay between BM and CRC diagnosis, n (%) | |
| Synchronous | 8 (10) |
| Metachronous | 74 (90) |
| ECM at BM diagnosis, n (%) | |
| No | 11 (14) |
| Yes | 70 (86) |
| Missing | 1 |
| Lung metastases at BM diagnosis, n (%) | |
| No | 23 (28) |
| Yes | 58 (72) |
| Missing | 1 |
| Liver metastases at BM diagnosis, n (%) | |
| No | 45 (56) |
| Yes | 36 (44) |
| Molecular Status | Primary Tumors (n = 82) | BM (n = 38) |
|---|---|---|
| KRAS status | ||
| Wild-type, n (%) | 35 (44) | 10 (26) |
| Mutant, n (%) | 44 (56) | 28 (74) |
| KRAS exon 2 at codon 12 | ||
| G12D | 14 (18) | 9 (23) |
| G12V | 14 (18) | 8 (21) |
| G12A | 5 (6) | 3 (8) |
| G12S | 3 (4) | 0 |
| G12C | 1 (1) | 1 (3) |
| G12R | 1 (1) | 1 (3) |
| KRAS exon 2 at codon 13 | ||
| G13D | 2 (3) | 3 (8) |
| G13R | 1 (1) | 1 (3) |
| KRAS exon 3 at codon 61 | 3 (4) | 2 (5) |
| KRAS exon 4 at codon 146 | 0 | 0 |
| Missing, n | 3 | 0 |
| NRAS status | ||
| Wild-type, n (%) | 74 (94) | 34 (89) |
| Mutant, n (%) | 5 (6) | 4 (11) |
| NRAS exon 2 at codon 12 or 13 | 1 (1) | 1 (3) |
| NRAS exon 3 at codon 61 | 4 (5) | 3 (8) |
| Missing, n | 3 | 0 |
| BRAF exon 15 at codon 600 | ||
| Wild-type, n (%) | 74 (94) | 36 (95) |
| Mutant, n (%) | 5 (6) | 2 (5) |
| Missing, n (%) | 3 | 0 |
| MMR status | ||
| pMMR, n (%) | 70 (95) | 36 (95) |
| dMMR, n (%) | 4 (5) | 2 (5) |
| Missing, n | 8 | 0 |
| cMET expression | ||
| Negative (0, 1+, 2+/3+ with FISH negative), n (%) | 76 (100) | 37 (100) |
| Positive (2+, 3+ with FISH positive), n (%) | 0 | 0 |
| Missing, n | 6 | 1 |
| HER-2 expression | ||
| Negative (0, 1+, 2+ with FISH negative), n (%) | 74 (99) | 37 (100) |
| Positive (2+ with FISH positive, 3+), n (%) | 1 (1) | 0 |
| Missing, n | 7 | 1 |
| ALK expression | ||
| Negative (0, 1+/2+/3+ with FISH negative), n (%) | 76 (100) | 37 (100) |
| Positive (1+/2+/3+ with FISH positive), n (%) | 0 | 0 |
| Missing, n (%) | 6 | 1 |
| ROS1 expression | ||
| Negative (0, 1+/2+/3+ with FISH negative), n (%) | 74 (100) | 37 (100) |
| Positive (1+/2+/3+ with FISH positive), n (%) | 0 | 0 |
| Missing, n | 8 | 1 |
| PD-1 expression | ||
| Negative, n (%) | 64 (86) | 37 (100) |
| Positive, n (%) | 10 (14) | 0 |
| Missing, n | 8 | 1 |
| PD-L1 expression | ||
| Negative, n (%) | 68 (93) | 35 (95) |
| Positive, n (%) | 5 (7) | 2 (5) |
| Missing, n | 9 | 1 |
| CD3 expression | ||
| Median rate, % (range) | 30 (0–80) | 11 (0–60) |
| Missing, n | 11 | 1 |
| CD8 expression | ||
| Median rate, % (range) | 11 (0–70) | 3 (0–50) |
| Missing, n | 7 | 2 |
| Brain Metastases | |||
|---|---|---|---|
| RAS status | |||
| Primary tumors | Wild-type | Mutant | Total |
| Wild-type, n (%) | 6 (17) | 3 (9) | 9 (26) |
| Mutant, n (%) | 0 | 26 (74) | 26 (74) |
| Total, n (%) | 6 (17) | 29 (83) | 35 |
| BRAF status | |||
| Primary tumors | Wild-type | Mutant | Total |
| Wild-type, n (%) | 33 (94) | 1 (3) | 34 (97) |
| Mutant, n (%) | 0 | 1 (3) | 1 (3) |
| Total, n (%) | 33 (94) | 2 (6) | 35 |
| MMR status | |||
| Primary tumors | pMMR | dMMR | Total |
| pMMR, n (%) | 30 (94) | 0 | 30 (94) |
| dMMR, n (%) | 0 | 2 (6) | 2 (6) |
| Total, n (%) | 30 (94) | 2 (6) | 32 |
| HER-2 expression | |||
| Primary tumors | Negative | Positive | Total |
| Negative, n (%) | 35 (100) | 0 | 35 (100) |
| Positive, n (%) | 0 | 0 | 0 (0) |
| Total, n (%) | 35 (100) | 0 (0) | 35 |
| cMET expression (IHC) | |||
| Primary tumors | Negative | Positive | Total |
| Negative, n (%) | 4 (13) | 7 (22) | 11 (34) |
| Positive, n (%) | 2 (6) | 19 (59) | 21 (66) |
| Total, n (%) | 6 (19) | 26 (81) | 32 |
| PD-1 expression | |||
| Primary tumors | Negative | Positive | Total |
| Negative, n (%) | 30 (94) | 0 | 30 (94) |
| Positive, n (%) | 2 (6) | 0 | 2 (6) |
| Total, n (%) | 32 (100) | 0 | 32 |
| PD-L1 expression | |||
| Primary tumors | Negative | Positive | Total |
| Negative, n (%) | 29 (91) | 2 (6) | 31 (97) |
| Positive, n (%) | 1 (3) | 0 | 1 (3) |
| Total, n (%) | 30 (94) | 2 (6) | 32 |
| CD3 expression | Primary tumor | Brain metastases | |
| Median rate, % (range) | 34 (0–80) | 15 (0–60) | |
| CD8 expression | Primary tumor | Brain metastases | |
| Median rate, % (range) | 10 (0–70) | 3 (0–50) | |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Variables | n | Median (Months) | p Value | HR | 95% CI | p Value |
| Gender (n = 82) | 0.79 * | 0.38 | ||||
| Male | 52 | 3.9 | 1 | |||
| Female | 30 | 4.3 | 0.8 | 0.5–1.4 | ||
| Age at BM diagnosis (n = 82) | 82 | 0.02 * | 1.0 | 1.0–1.0 | 0.62 | |
| Site of primary tumor (n = 82) | 0.23 | |||||
| Ascending colon | 20 | 4.5 | ||||
| Descending colon | 24 | 5.9 | ||||
| Rectum | 35 | 2.9 | ||||
| Tumor grade (n = 70) | 0.05 | |||||
| Well or moderately differentiated | 61 | 3.9 | ||||
| Poorly differentiated | 9 | 4.6 | ||||
| RAS status (n = 79) | 0.65 | |||||
| Wild-type | 30 | 3.6 | ||||
| Mutant | 49 | 4.3 | ||||
| BRAF status (n = 79) | 0.03 * | 0.76 | ||||
| Wild-type | 74 | 4.2 | 1 | |||
| Mutant | 5 | 3.3 | 1.2 | 0.3–4.2 | ||
| MMR status (n = 74) | 0.68 | |||||
| pMMR | 70 | 4.1 | ||||
| dMMR | 4 | 4.0 | ||||
| PD-1 expression (n = 74) | 0.79 | |||||
| Negative | 64 | 4.2 | ||||
| Positive | 10 | 3.6 | ||||
| PD-L1 expression (n = 73) | 0.009 * | 0.02 | ||||
| Negative | 68 | 4.2 | 1 | |||
| Positive | 5 | 1.8 | 5.0 | 1.4–18.5 | ||
| CD3 expression (n = 71) | 71 | 0.08 | ||||
| CD8 expression (n = 75) | 75 | 0.45 | ||||
| ECOG performance status (n = 79) | 0.0003 * | 0.07 | ||||
| <2 | 43 | 7.3 | 1 | |||
| ≥2 | 36 | 3.2 | 1.8 | 1.0–3.4 | ||
| Number of BM (n = 82) | 0.003 * | 0.01 | ||||
| Single | 43 | 6.3 | 1 | |||
| Multiple | 39 | 3.1 | 2.0 | 1.2–3.4 | ||
| Lung metastases at BM diagnosis (n = 81) | 0.0003 * | 0.005 | ||||
| No | 23 | 11.7 | 1 | |||
| Yes | 58 | 3.6 | 2.5 | 1.3–4.8 | ||
| Liver metastases at BM diagnosis (n = 81) | ||||||
| No | 45 | 4.3 | 0.31 | |||
| Yes | 36 | 3.7 | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roussille, P.; Tachon, G.; Villalva, C.; Milin, S.; Frouin, E.; Godet, J.; Berger, A.; Emambux, S.; Petropoulos, C.; Wager, M.; et al. Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases. Cancers 2018, 10, 504. https://doi.org/10.3390/cancers10120504
Roussille P, Tachon G, Villalva C, Milin S, Frouin E, Godet J, Berger A, Emambux S, Petropoulos C, Wager M, et al. Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases. Cancers. 2018; 10(12):504. https://doi.org/10.3390/cancers10120504
Chicago/Turabian StyleRoussille, Pauline, Gaelle Tachon, Claire Villalva, Serge Milin, Eric Frouin, Julie Godet, Antoine Berger, Sheik Emambux, Christos Petropoulos, Michel Wager, and et al. 2018. "Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases" Cancers 10, no. 12: 504. https://doi.org/10.3390/cancers10120504
APA StyleRoussille, P., Tachon, G., Villalva, C., Milin, S., Frouin, E., Godet, J., Berger, A., Emambux, S., Petropoulos, C., Wager, M., Karayan-Tapon, L., & Tougeron, D. (2018). Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases. Cancers, 10(12), 504. https://doi.org/10.3390/cancers10120504





