A Cell-Autonomous Oncosuppressive Role of Human RNASET2 Affecting ECM-Mediated Oncogenic Signaling
Abstract
:1. Introduction
2. Results
2.1. RNASET2 Transcript Expression in EOC Is Associated to Better Prognosis
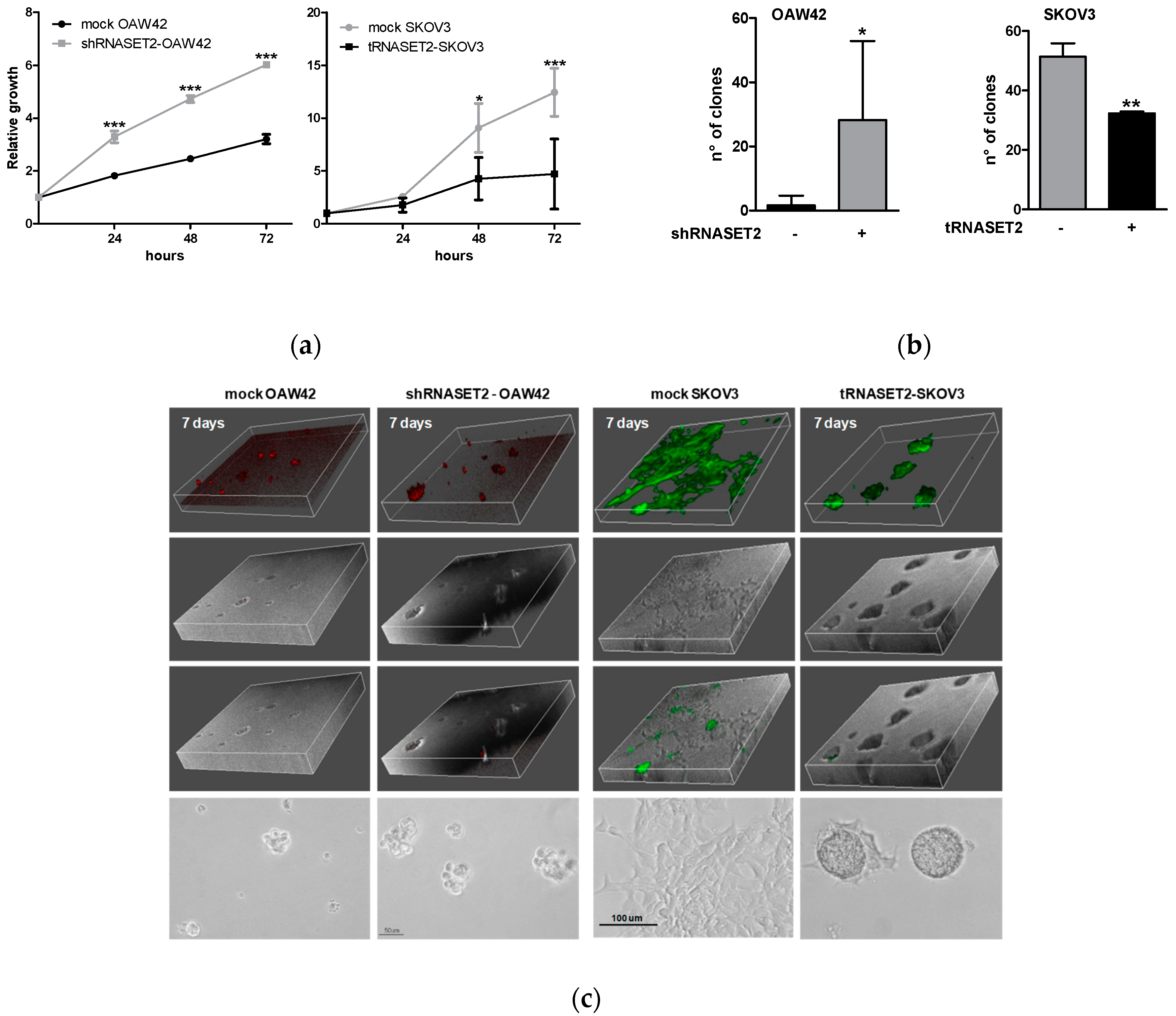
2.2. RNASET2 Depletion Causes Phenotypic Changes in EOC Cellular Models
2.3. RNASET2 Expression Negatively Impinges EOC Cell Growth
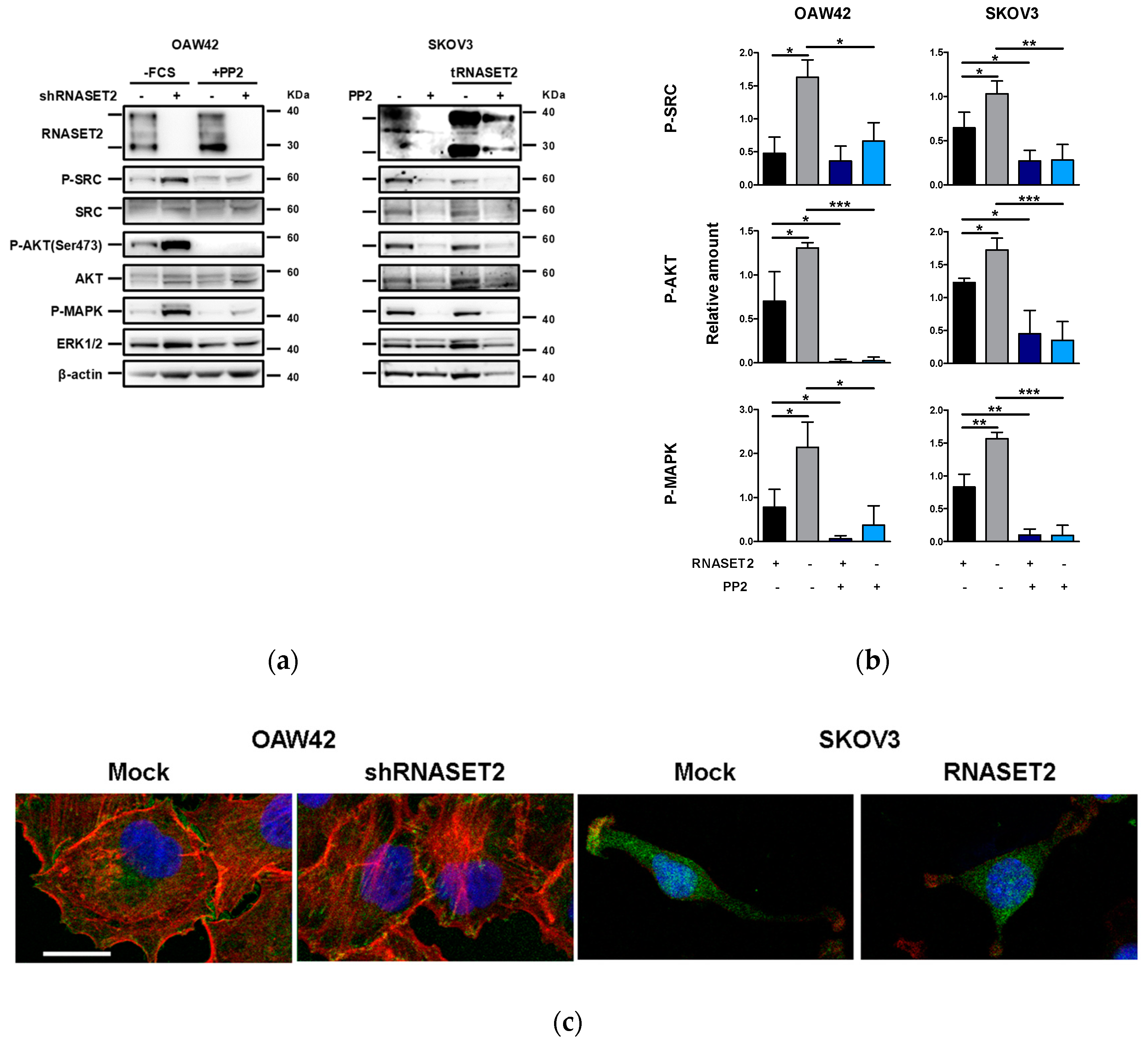
2.4. ‘RNASET2 Phenotype’ Negatively Modulates Src Activation
2.5. Src Inhibitor Restores ‘RNASET2 Phenotype’
2.6. EOC Cells with ‘RNASET2 Phenotype’ Are Sensitive to Src Inhibitor
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Plasmids
4.2. Patient Samples
4.3. In Silico Analysis of RNASET2 Expression and Survival Analysis
4.4. IHC
4.5. Antibodies
4.6. RNA Extraction and Quantitative Real-Time RT–PCR Analysis
4.7. Western Blotting
4.8. Immunofluorescence (IF) and Confocal Microscopy
4.9. Cell Proliferation Assay of 2D and 3D Cultures
4.10. Soft Agar Assay
4.11. Growth and Invasion on Matrigel®
4.12. FACS Analysis
4.13. Adhesion Assay and Treatment with Inhibitors
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coleman, R.L.; Monk, B.J.; Sood, A.K.; Herzog, T.J. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat. Rev. Clin. Oncol. 2013, 10, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashashati, A.; Ha, G.; Tone, A.; Ding, J.; Prentice, L.M.; Roth, A.; Rosner, J.; Shumansky, K.; Kalloger, S.; Senz, J.; et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol. 2013, 231, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbolina, M.V. Molecular Mechanisms Regulating Organ-Specific Metastases in Epithelial Ovarian Carcinoma. Cancers 2018, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Campomenosi, P.; Salis, S.; Lindqvist, C.; Mariani, D.; Nordstrom, T.; Acquati, F.; Taramelli, R. Characterization of RNASET2, the first human member of the Rh/T2/S family of glycoproteins. Arch. Biochem. Biophys. 2006, 449, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Vidalino, L.; Monti, L.; Haase, A.; Moro, A.; Acquati, F.; Taramelli, R.; Macchi, P. Intracellular trafficking of RNASET2, a novel component of P-bodies. Biol. Cell 2012, 104, 13–21. [Google Scholar] [CrossRef]
- Acquati, F.; Morelli, C.; Cinquetti, R.; Bianchi, M.G.; Porrini, D.; Varesco, L.; Gismondi, V.; Rocchetti, R.; Talevi, S.; Possati, L.; et al. Cloning and characterization of a senescence inducing and class II tumor suppressor gene in ovarian carcinoma at chromosome region 6q27. Oncogene 2001, 20, 980–988. [Google Scholar] [CrossRef] [Green Version]
- Acquati, F.; Possati, L.; Ferrante, L.; Campomenosi, P.; Talevi, S.; Bardelli, S.; Margiotta, C.; Russo, A.; Bortoletto, E.; Rocchetti, R.; et al. Tumor and metastasis suppression by the human RNASET2 gene. Int. J. Oncol. 2005, 26, 1159–1168. [Google Scholar] [CrossRef]
- Acquati, F.; Lualdi, M.; Bertilaccio, S.; Monti, L.; Turconi, G.; Fabbri, M.; Grimaldi, A.; Anselmo, A.; Inforzato, A.; Collotta, A.; et al. Loss of function of Ribonuclease T2, an ancient and phylogenetically conserved RNase, plays a crucial role in ovarian tumorigenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 8140–8145. [Google Scholar] [CrossRef] [Green Version]
- Lualdi, M.; Pedrini, E.; Rea, K.; Monti, L.; Scaldaferri, D.; Gariboldi, M.; Camporeale, A.; Ghia, P.; Monti, E.; Tomassetti, A.; et al. Pleiotropic modes of action in tumor cells of RNASET2, an evolutionary highly conserved extracellular RNase. Oncotarget 2015, 6, 7851–7865. [Google Scholar] [CrossRef] [Green Version]
- Scaldaferri, D.; Bosi, A.; Fabbri, M.; Pedrini, E.; Inforzato, A.; Valli, R.; Frattini, A.; De Vito, A.; Noonan, D.M.; Taramelli, R.; et al. The human RNASET2 protein affects the polarization pattern of human macrophages in vitro. Immunol. Lett. 2018, 203, 102–111. [Google Scholar] [CrossRef]
- Roggiani, F.; Mezzanzanica, D.; Rea, K.; Tomassetti, A. Guidance of Signaling Activations by Cadherins and Integrins in Epithelial Ovarian Cancer Cells. Int. J. Mol. Sci. 2016, 17, 1387. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, P.; Roiz, L.; Angelkovitch, B.; Schwartz, B.; Shoseyov, O. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics: Expression, purification, and characterization. Cancer 2006, 107, 2760–2769. [Google Scholar] [CrossRef] [PubMed]
- Monti, L.; Rodolfo, M.; Lo, R.G.; Noonan, D.; Acquati, F.; Taramelli, R. RNASET2 as a tumor antagonizing gene in a melanoma cancer model. Oncol. Res. 2008, 17, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Acquati, F.; Bertilaccio, S.; Grimaldi, A.; Monti, L.; Cinquetti, R.; Bonetti, P.; Lualdi, M.; Vidalino, L.; Fabbri, M.; Sacco, M.G.; et al. Microenvironmental control of malignancy exerted by RNASET2, a widely conserved extracellular RNase. Proc. Natl. Acad. Sci. USA 2011, 108, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.-P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65–74. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Hernandez, L.; Kim, M.K.; Lyle, L.T.; Bunch, K.P.; House, C.D.; Ning, F.; Noonan, A.M.; Annunziata, C.M. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol. Oncol. 2016, 142, 332–340. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, J.; Horiuchi, A.; Kikuchi, N.; Hayashi, A.; Osada, R.; Ohira, S.; Shiozawa, T.; Konishi, I. Changes in the expression of E-cadherin repressors, Snail, Slug, SIP1, and Twist, in the development and progression of ovarian carcinoma: The important role of Snail in ovarian tumorigenesis and progression. Med. Mol. Morphol. 2009, 42, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Roggiani, F.; Tomassetti, A. Fondazione IRCCS Istituto nazionale dei Tumori, Milan, Italy. Unpublished data. 2017. [Google Scholar]
- Rea, K.; Roggiani, F.; De Cecco, L.; Raspagliesi, F.; Carcangiu, M.L.; Nair-Menon, J.; Bagnoli, M.; Bortolomai, I.; Mezzanzanica, D.; Canevari, S.; et al. Simultaneous E-cadherin and PLEKHA7 expression negatively affects E-cadherin/EGFR mediated ovarian cancer cell growth. J. Exp. Clin. Cancer Res. 2018, 37, 146. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, B.; Shoseyov, O.; Melnikova, V.O.; McCarty, M.; Leslie, M.; Roiz, L.; Smirnoff, P.; Hu, G.F.; Lev, D.; Bar-Eli, M. ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis, and metastasis. Cancer Res. 2007, 67, 5258–5266. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, M.; Wu, J.; Ma, Y.; Li, T.; Chen, Q.; Zhang, X.; Xiang, L. Stress-induced RNASET2 overexpression mediates melanocyte apoptosis via the TRAF2 pathway in vitro. Cell Death Dis. 2014, 5, e1022. [Google Scholar] [CrossRef]
- Caputa, G.; Zhao, S.; Criado, A.E.; Ory, D.S.; Duncan, J.G.; Schaffer, J.E. RNASET2 is required for ROS propagation during oxidative stress-mediated cell death. Cell Death Differ. 2016, 23, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Roiz, L.; Smirnoff, P.; Lewin, I.; Shoseyov, O.; Schwartz, B. Human recombinant RNASET2: A potential anti-cancer drug. Oncoscience 2016, 3, 71–84. [Google Scholar] [PubMed]
- Wang, Q.; Wang, X.; Xiang, L. Role and Mechanism of RNASET2 in the Pathogenesis of Vitiligo. J. Investig. Dermatol. Symp. Proc. 2015, 17, 48–50. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Caveolae and signalling in cancer. Nat. Rev. Cancer 2015, 15, 225–237. [Google Scholar] [CrossRef]
- Steinfelder, S.; Andersen, J.F.; Cannons, J.L.; Feng, C.G.; Joshi, M.; Dwyer, D.; Caspar, P.; Schwartzberg, P.L.; Sher, A.; Jankovic, D. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1). J. Exp. Med. 2009, 206, 1681–1690. [Google Scholar] [CrossRef] [Green Version]
- Aleshin, A.; Finn, R.S. SRC: A century of science brought to the clinic. Neoplasia 2010, 12, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Chen, C.; Xu, M.M.; Li, J.D.; Xiao, J.; Zhu, X.F. Expression of c-Src and phospho-Src in epithelial ovarian carcinoma. Mol. Cell Biochem. 2013, 376, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.W.; McLean, G.W.; Wyke, A.W.; Paraskeva, C.; Parkinson, E.K.; Frame, M.C.; Brunton, V.G. The catalytic activity of the Src family kinases is required to disrupt cadherin-dependent cell-cell contacts. Mol. Biol. Cell 2000, 11, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Miotti, S.; Tomassetti, A.; Facetti, I.; Sanna, E.; Berno, V.; Canevari, S. Simultaneous Expression of Caveolin-1 and E-Cadherin in Ovarian Carcinoma Cells Stabilizes Adherens Junctions through Inhibition of src-Related Kinases. Am. J. Pathol. 2005, 167, 1411–1427. [Google Scholar] [CrossRef] [Green Version]
- Han, L.Y.; Landen, C.N.; Trevino, J.G.; Halder, J.; Lin, Y.G.; Kamat, A.A.; Kim, T.J.; Merritt, W.M.; Coleman, R.L.; Gershenson, D.M.; et al. Antiangiogenic and antitumor effects of SRC inhibition in ovarian carcinoma. Cancer Res. 2006, 66, 8633–8639. [Google Scholar] [CrossRef]
- Konecny, G.E.; Glas, R.; Dering, J.; Manivong, K.; Qi, J.; Finn, R.S.; Yang, G.R.; Hong, K.L.; Ginther, C.; Winterhoff, B.; et al. Activity of the multikinase inhibitor dasatinib against ovarian cancer cells. Br. J. Cancer 2009, 101, 1699–1708. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Fu, S. An overview of tyrosine kinase inhibitors for the treatment of epithelial ovarian cancer. Expert Opin. Investig. Drugs 2016, 25, 15–30. [Google Scholar] [CrossRef]
- Ricci, F.; Bizzaro, F.; Cesca, M.; Guffanti, F.; Ganzinelli, M.; Decio, A.; Ghilardi, C.; Perego, P.; Fruscio, R.; Buda, A.; et al. Patient-derived ovarian tumor xenografts recapitulate human clinicopathology and genetic alterations. Cancer Res. 2014, 74, 6980–6990. [Google Scholar] [CrossRef]
- Ledermann, J.A. Front-line therapy of advanced ovarian cancer: New approaches. Ann. Oncol 2017, 28, viii46–viii50. [Google Scholar] [CrossRef]
- Garcia-Roman, J.; Zentella-Dehesa, A. Vascular permeability changes involved in tumor metastasis. Cancer Lett. 2013, 335, 259–269. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lanczky, A.; Szallasi, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberti, C.; Pinciroli, P.; Valeri, B.; Ferri, R.; Ditto, A.; Umezawa, K.; Sensi, M.; Canevari, S.; Tomassetti, A. Ligand-dependent EGFR activation induces the co-expression of IL-6 and PAI-1 via the NFkB pathway in advanced-stage epithelial ovarian cancer. Oncogene 2012, 31, 4139–4149. [Google Scholar] [CrossRef] [PubMed]
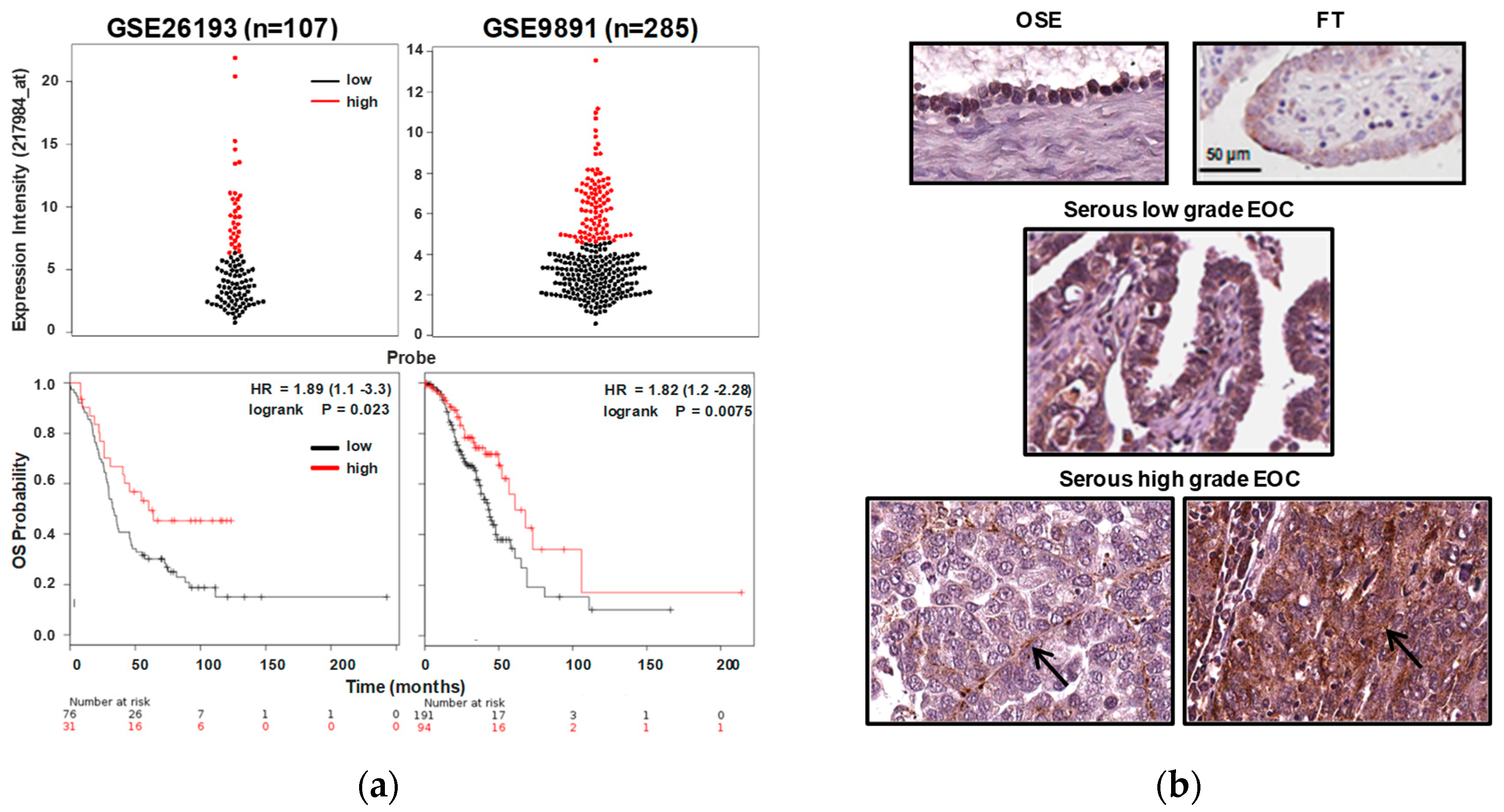




| Histological Subtype | RNASET2 Expression | |||
|---|---|---|---|---|
| Negative (27.5%) | Positive (72.5%) | |||
| Score ** 0 (n = 27) | Score 1 (n = 43) | Score 2 (n = 32) | ||
| OSE * | - | - | 1 | |
| FTE * | - | 1 | - | |
| Type I (n = 47) | Endometrioid | 0 | 2 | 2 |
| Serous | 4 | 10 | 3 | |
| Mucinous | 4 | 9 | 7 | |
| Clear cell | 0 | 3 | 3 | |
| Type II (n = 54) | Endometrioid | 2 | 4 | 0 |
| Serous | 13 | 16 | 16 | |
| Undifferentiated | 3 | 0 | 0 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roggiani, F.; Riva, C.; Raspagliesi, F.; Porta, G.; Valli, R.; Taramelli, R.; Acquati, F.; Mezzanzanica, D.; Tomassetti, A. A Cell-Autonomous Oncosuppressive Role of Human RNASET2 Affecting ECM-Mediated Oncogenic Signaling. Cancers 2019, 11, 255. https://doi.org/10.3390/cancers11020255
Roggiani F, Riva C, Raspagliesi F, Porta G, Valli R, Taramelli R, Acquati F, Mezzanzanica D, Tomassetti A. A Cell-Autonomous Oncosuppressive Role of Human RNASET2 Affecting ECM-Mediated Oncogenic Signaling. Cancers. 2019; 11(2):255. https://doi.org/10.3390/cancers11020255
Chicago/Turabian StyleRoggiani, Francesca, Cristina Riva, Francesco Raspagliesi, Giovanni Porta, Roberto Valli, Roberto Taramelli, Francesco Acquati, Delia Mezzanzanica, and Antonella Tomassetti. 2019. "A Cell-Autonomous Oncosuppressive Role of Human RNASET2 Affecting ECM-Mediated Oncogenic Signaling" Cancers 11, no. 2: 255. https://doi.org/10.3390/cancers11020255





