Regulation of KIF2A by Antitumor miR-451a Inhibits Cancer Cell Aggressiveness Features in Lung Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
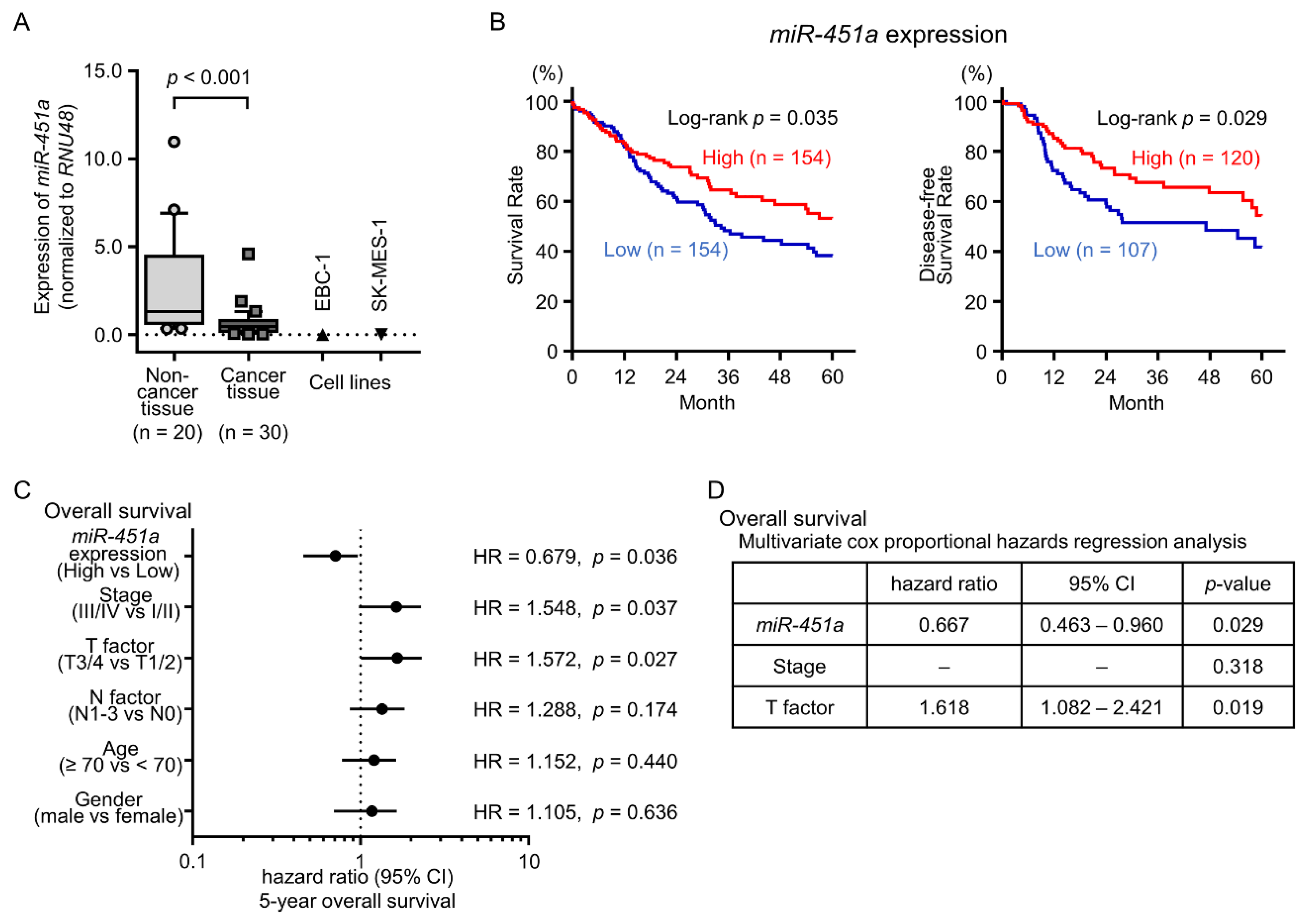
2.1. Downregulation of miR-451a in LUSQ Clinical Specimens and Its Clinical Significance
2.2. Induction of Apoptotic Cells by Ectopic Expression of miR-451a in LUSQ Cells
2.3. Effects of Ectopic Expression of miR-451a on LUSQ Cell Migration and Invasion
2.4. Screening of Putative Target Genes by miR-451a Regulation in LUSQ Cells
2.5. Expression of KIF2A Was Directly Controlled by miR-451a in LUSQ Cells
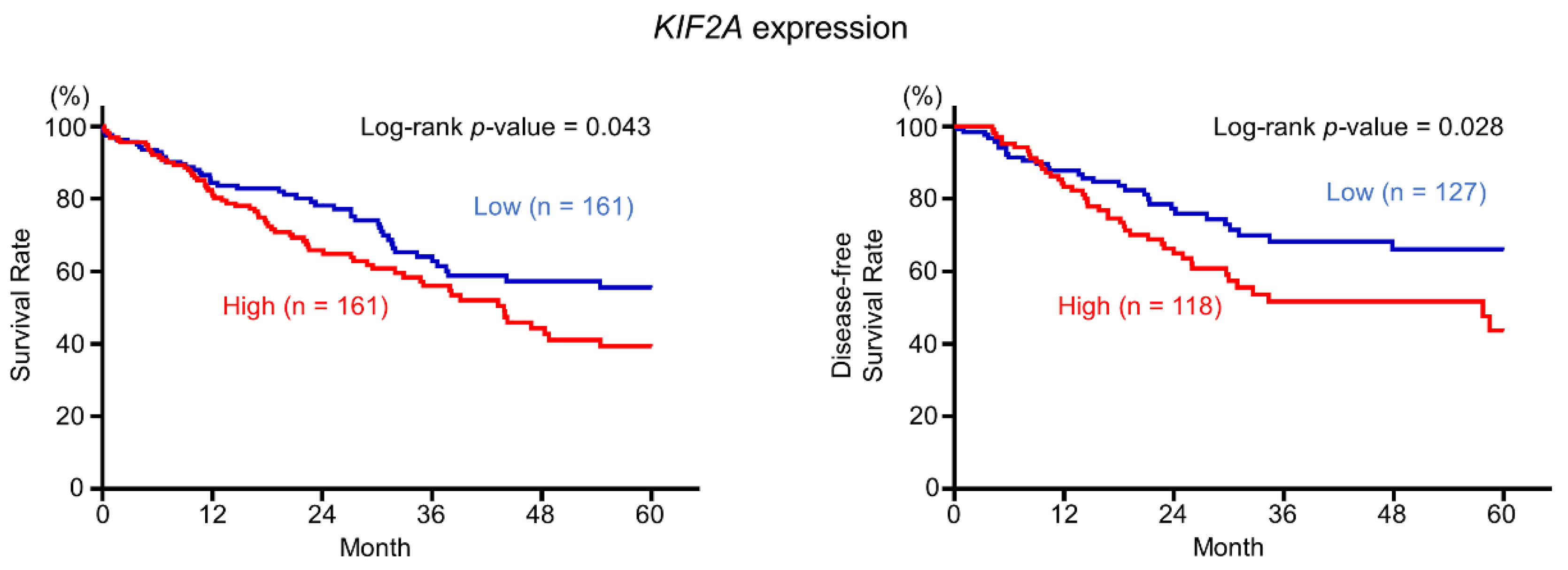
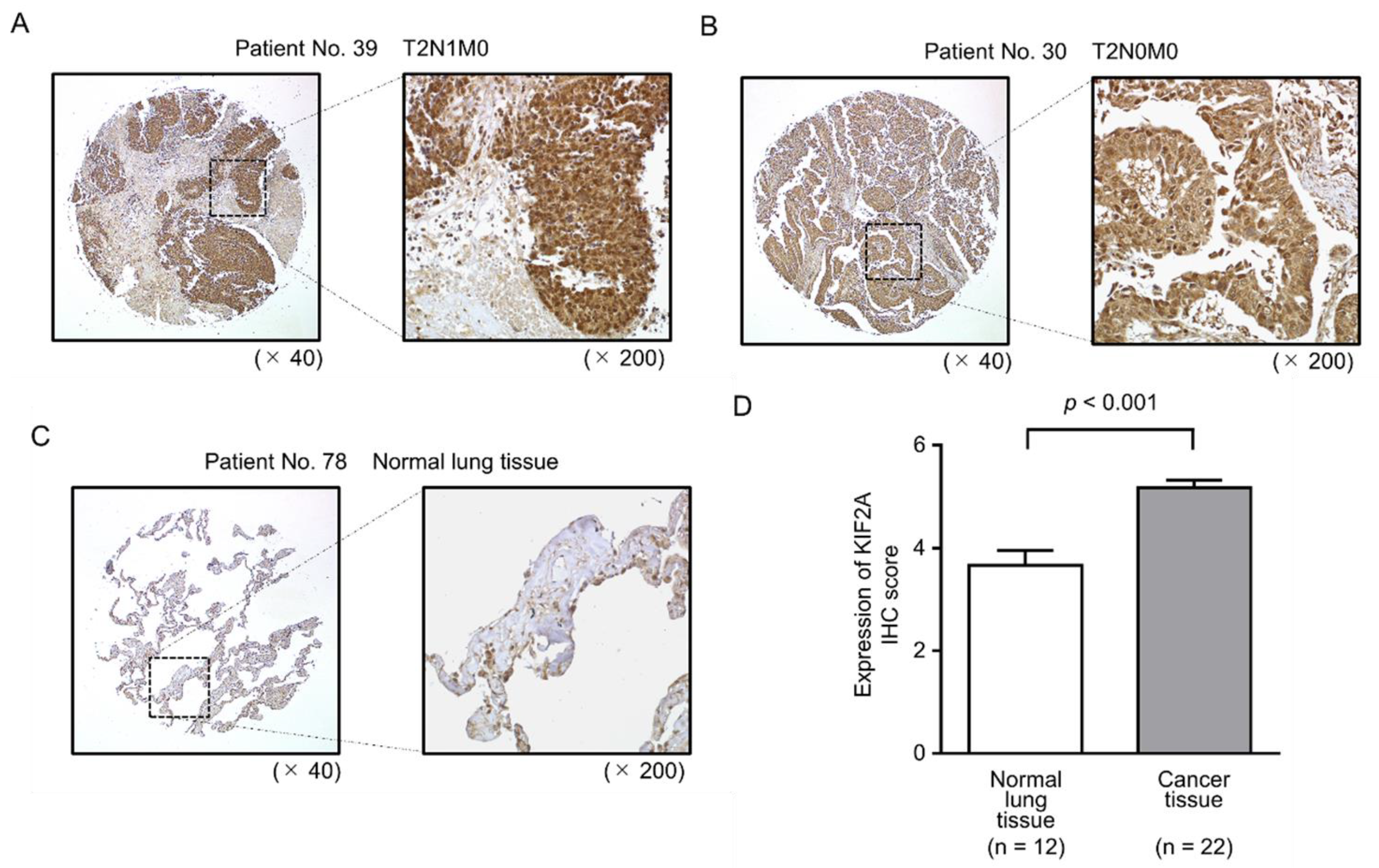
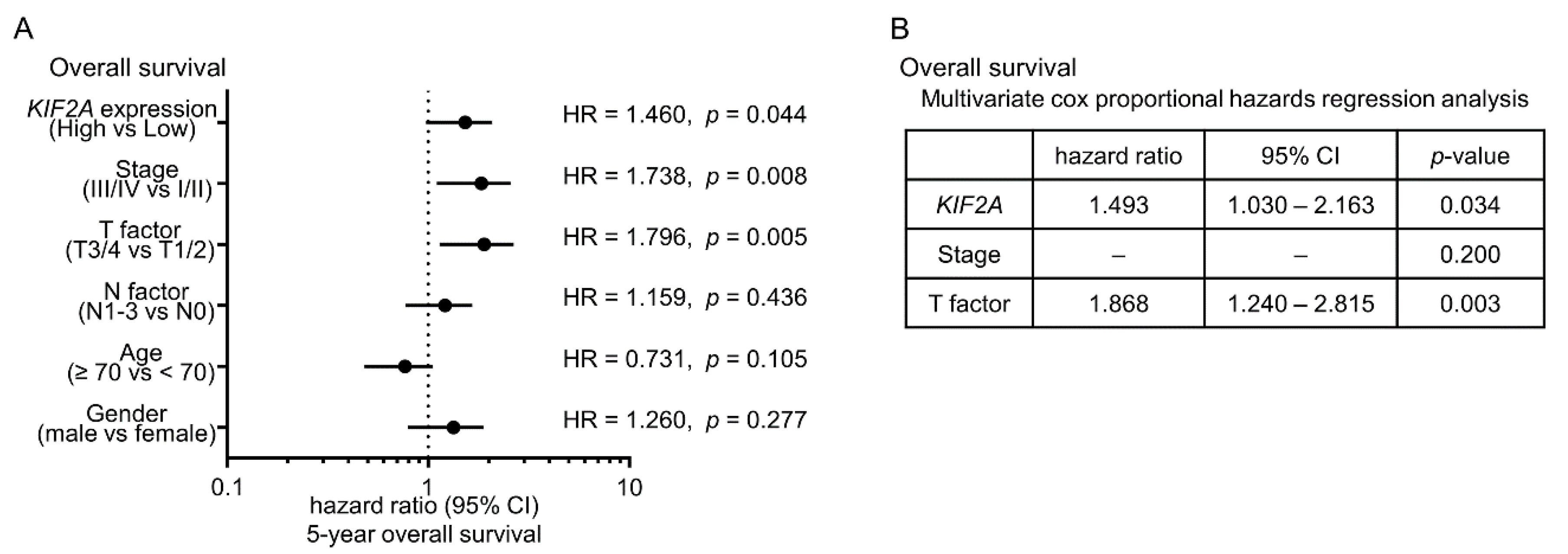
2.6. Aberrant Expression of KIF2A and Its Clinical Significance in LUSQ
2.7. Effects of KIF2A Knockdown on Cell Proliferation and Induction of Apoptotic Cells in LUSQ Cells
2.8. Effects of KIF2A Silencing on Cancer Cell Migration and Invasion in LUSQ Cells
2.9. Identification of KIF2A-Mediated Downstream Pathways in LUSQ Cells
3. Discussion
4. Materials and Methods
4.1. Clinical Samples Collection, Cell Lines, and RNA Extraction
4.2. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.3. Transfections with Mature miRNA and siRNA into LUSQ Cell Lines
4.4. Cell proliferation, Migration, and Invasion Assays
4.5. Apoptosis Assays
4.6. Identification of Putative Target Genes Regulated by miR-451a in LUSQ Cells
4.7. Clinical Database Analysis
4.8. Plasmid Construction and Dual-Luciferase Reporter Assay
4.9. Western Blotting and Immunohistochemistry
4.10. Identification of Downstream Targets Regulated by KIF2A in LUSQ Cells
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar]
- Travis, W.D. Pathology of lung cancer. Clin. Chest Med. 2011, 32, 669–692. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann. Oncol. 2015, 26, 1877–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.I.; Perol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive non-small-cell lung cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Hammerman, P.S.; Sos, M.L.; Lara, P.N., Jr.; Hirsch, F.R. Squamous cell lung cancer: From tumor genomics to cancer therapeutics. Clin. Cancer Res. 2015, 21, 2236–2243. [Google Scholar] [CrossRef] [PubMed]
- Little, A.G.; Gay, E.G.; Gaspar, L.E.; Stewart, A.K. National survey of non-small cell lung cancer in the United States: Epidemiology, pathology and patterns of care. Lung Cancer 2007, 57, 253–260. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Baranwal, S.; Alahari, S.K. miRNA control of tumor cell invasion and metastasis. Int. J. Cancer 2010, 126, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Mataki, H.; Seki, N.; Kumamoto, T.; Kamikawaji, K.; Inoue, H. MicroRNAs in non-small cell lung cancer and idiopathic pulmonary fibrosis. J. Hum. Genet. 2017, 62, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016, 7, 72084–72098. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, T.; Koshizuka, K.; Seki, N.; Mizuno, K.; Okato, A.; Arai, T.; Misono, S.; Uchida, A.; Kumamoto, T.; Inoue, H. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int. J. Oncol. 2018, 52, 913–924. [Google Scholar] [CrossRef]
- Mizuno, K.; Seki, N.; Mataki, H.; Matsushita, R.; Kamikawaji, K.; Kumamoto, T.; Takagi, K.; Goto, Y.; Nishikawa, R.; Kato, M.; et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int. J. Oncol. 2016, 48, 450–460. [Google Scholar] [CrossRef]
- Kumamoto, T.; Seki, N.; Mataki, H.; Mizuno, K.; Kamikawaji, K.; Samukawa, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Regulation of TPD52 by antitumor microRNA-218 suppresses cancer cell migration and invasion in lung squamous cell carcinoma. Int. J. Oncol. 2016, 49, 1870–1880. [Google Scholar] [CrossRef]
- Uchida, A.; Seki, N.; Mizuno, K.; Misono, S.; Yamada, Y.; Kikkawa, N.; Sanada, H.; Kumamoto, T.; Suetsugu, T.; Inoue, H. Involvement of dual-strand of the miR-144 duplex and their targets in the pathogenesis of lung squamous cell carcinoma. Cancer Sci. 2018. [Google Scholar] [CrossRef]
- Mataki, H.; Enokida, H.; Chiyomaru, T.; Mizuno, K.; Matsushita, R.; Goto, Y.; Nishikawa, R.; Higashimoto, I.; Samukawa, T.; Nakagawa, M.; et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J. Hum. Genet. 2015, 60, 53–61. [Google Scholar] [CrossRef]
- Kojima, S.; Chiyomaru, T.; Kawakami, K.; Yoshino, H.; Enokida, H.; Nohata, N.; Fuse, M.; Ichikawa, T.; Naya, Y.; Nakagawa, M.; et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br. J. Cancer 2012, 106, 405–413. [Google Scholar] [CrossRef]
- Nohata, N.; Hanazawa, T.; Kikkawa, N.; Sakurai, D.; Sasaki, K.; Chiyomaru, T.; Kawakami, K.; Yoshino, H.; Enokida, H.; Nakagawa, M.; et al. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2011, 39, 1099–1107. [Google Scholar]
- Nohata, N.; Hanazawa, T.; Enokida, H.; Seki, N. microRNA-1/133a and microRNA-206/133b clusters: Dysregulation and functional roles in human cancers. Oncotarget 2012, 3, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Enokida, H.; Chiyomaru, T.; Kinoshita, T.; Nakagawa, M.; Naya, Y.; Ichikawa, T.; Seki, N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 2014, 5, 7748–7759. [Google Scholar] [CrossRef]
- Yoshino, H.; Enokida, H.; Itesako, T.; Kojima, S.; Kinoshita, T.; Tatarano, S.; Chiyomaru, T.; Nakagawa, M.; Seki, N. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013, 104, 1567–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Enokida, H.; Yoshino, H.; Itesako, T.; Chiyomaru, T.; Kinoshita, T.; Fuse, M.; Nishikawa, R.; Goto, Y.; Naya, Y.; et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J. Hum. Genet. 2014, 59, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Kurozumi, A.; Kato, M.; Enokida, H.; Matsushita, R.; Yamazaki, K.; Ishida, Y.; Nakagawa, M.; et al. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 2015, 113, 1055–1065. [Google Scholar] [CrossRef]
- Fukumoto, I.; Kinoshita, T.; Hanazawa, T.; Kikkawa, N.; Chiyomaru, T.; Enokida, H.; Yamamoto, N.; Goto, Y.; Nishikawa, R.; Nakagawa, M.; et al. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br. J. Cancer 2014, 111, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Yamada, Y.; Arai, T.; Kojima, S.; Sugawara, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018. [Google Scholar] [CrossRef]
- Yamada, Y.; Arai, T.; Sugawara, S.; Okato, A.; Kato, M.; Kojima, S.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Impact of novel oncogenic pathways regulated by antitumor miR-451a in renal cell carcinoma. Cancer Sci. 2018, 109, 1239–1253. [Google Scholar] [CrossRef]
- Matsushita, R.; Seki, N.; Chiyomaru, T.; Inoguchi, S.; Ishihara, T.; Goto, Y.; Nishikawa, R.; Mataki, H.; Tatarano, S.; Itesako, T.; et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br. J. Cancer 2015, 113, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Bandres, E.; Bitarte, N.; Arias, F.; Agorreta, J.; Fortes, P.; Agirre, X.; Zarate, R.; Diaz-Gonzalez, J.A.; Ramirez, N.; Sola, J.J.; et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009, 15, 2281–2290. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, H.; Powathil, G.; Kim, H.; Trucu, D.; Lee, W.; Lawler, S.; Chaplain, M. Role of extracellular matrix and microenvironment in regulation of tumor growth and LAR-mediated invasion in glioblastoma. PLoS ONE 2018, 13, e0204865. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, N.; Guo, R.; Jiang, W.; He, Q.M.; Xu, Y.F.; Li, Y.Q.; Tang, L.L.; Mao, Y.P.; Sun, Y.; et al. MiR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol. Cancer 2013, 12, 123. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.Q.; Yu, C.; Zhang, X.F.; Liu, Z.F.; Wang, R.; Jiang, M.; Chen, H.; Yan, F.; Tao, M.; Chen, L.B.; et al. HDAC3-mediated silencing of miR-451 decreases chemosensitivity of patients with metastatic castration-resistant prostate cancer by targeting NEDD9. Ther. Adv. Med. Oncol. 2018, 10, 1758835918783132. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Z.X.; Yang, J.S.; Pan, X.; De, W.; Chen, L.B. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting Ras-related protein 14 (RAB14). Oncogene 2011, 30, 2644–2658. [Google Scholar] [CrossRef]
- Goto, A.; Tanaka, M.; Yoshida, M.; Umakoshi, M.; Nanjo, H.; Shiraishi, K.; Saito, M.; Kohno, T.; Kuriyama, S.; Konno, H.; et al. The low expression of miR-451 predicts a worse prognosis in non-small cell lung cancer cases. PLoS ONE 2017, 12, e0181270. [Google Scholar] [CrossRef]
- Chang, C.; Liu, J.; He, W.; Qu, M.; Huang, X.; Deng, Y.; Shen, L.; Zhao, X.; Guo, H.; Jiang, J.; et al. A regulatory circuit HP1gamma/miR-451a/c-Myc promotes prostate cancer progression. Oncogene 2018, 37, 415–426. [Google Scholar] [CrossRef]
- Desai, A.; Verma, S.; Mitchison, T.J.; Walczak, C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell 1999, 96, 69–78. [Google Scholar] [CrossRef]
- Ganem, N.J.; Compton, D.A. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004, 166, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Ems-McClung, S.C.; Walczak, C.E. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 2010, 21, 276–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Ma, S.; Ma, R.; Qu, X.; Liu, W.; Lv, C.; Zhao, S.; Gong, Y. KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer 2014, 14, 461. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Qu, X.; Zhang, X.Y.; Zhou, C.J.; Liu, G.X.; Dong, Z.Q.; Wei, F.C.; Sun, S.Z. Overexpression of Kif2a promotes the progression and metastasis of squamous cell carcinoma of the oral tongue. Oral Oncol. 2010, 46, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Wang, X.; Zhu, H.; Wang, W.; Zhang, S.; Wang, Z. KIF2A overexpression and its association with clinicopathologic characteristics and unfavorable prognosis in colorectal cancer. Tumour Biol. 2015, 36, 8895–8902. [Google Scholar] [CrossRef] [PubMed]
- Sheng, N.; Xu, Y.Z.; Xi, Q.H.; Jiang, H.Y.; Wang, C.Y.; Zhang, Y.; Ye, Q. Overexpression of KIF2A is Suppressed by miR-206 and Associated with Poor Prognosis in Ovarian Cancer. Cell. Physiol. Biochem. 2018, 50, 810–822. [Google Scholar] [CrossRef]
- Xie, T.; Li, X.; Ye, F.; Lu, C.; Huang, H.; Wang, F.; Cao, X.; Zhong, C. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2018, 497, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, X.; Shan, C.; Li, Y.; Li, F. Minichromosome maintenance protein 2 correlates with the malignant status and regulates proliferation and cell cycle in lung squamous cell carcinoma. Onco. Targets Ther. 2018, 11, 5025–5034. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Kinoshita, I.; Shimizu, Y.; Kikuchi, E.; Takeda, K.; Aburatani, H.; Oizumi, S.; Konishi, J.; Kaga, K.; Matsuno, Y.; et al. Minichromosome maintenance (MCM) protein 4 as a marker for proliferation and its clinical and clinicopathological significance in non-small cell lung cancer. Lung Cancer 2011, 72, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Mirsadraee, S.; Oswal, D.; Alizadeh, Y.; Caulo, A.; van Beek, E., Jr. The 7th lung cancer TNM classification and staging system: Review of the changes and implications. World J. Radiol. 2012, 4, 128–134. [Google Scholar] [CrossRef]
- Mataki, H.; Seki, N.; Chiyomaru, T.; Enokida, H.; Goto, Y.; Kumamoto, T.; Machida, K.; Mizuno, K.; Nakagawa, M.; Inoue, H. Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. Int. J. Oncol. 2015, 46, 1039–1050. [Google Scholar] [CrossRef]
- Mizuno, K.; Mataki, H.; Arai, T.; Okato, A.; Kamikawaji, K.; Kumamoto, T.; Hiraki, T.; Hatanaka, K.; Inoue, H.; Seki, N. The microRNA expression signature of small cell lung cancer: Tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J. Hum. Genet. 2017, 62, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Kojima, S.; Yamada, Y.; Sugawara, S.; Kato, M.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Pirin: A potential novel therapeutic target for castration-resistant prostate cancer regulated by miR-455-5p. Mol. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, R.; Yoshino, H.; Enokida, H.; Goto, Y.; Miyamoto, K.; Yonemori, M.; Inoguchi, S.; Nakagawa, M.; Seki, N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget 2016, 7, 28460–28487. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Misono, S.; Seki, N.; Mizuno, K.; Yamada, Y.; Uchida, A.; Arai, T.; Kumamoto, T.; Sanada, H.; Suetsugu, T.; Inoue, H. Dual strands of the miR-145 duplex (miR-145-5p and miR-145-3p) regulate oncogenes in lung adenocarcinoma pathogenesis. J. Hum. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Sugawara, S.; Arai, T.; Kojima, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Molecular pathogenesis of renal cell carcinoma: Impact of the anti-tumor miR-29 family on gene regulation. Int. J. Urol. 2018, 25, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, K.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Fukumoto, I.; Katada, K.; Okamoto, Y.; Seki, N. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: Dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget 2017, 8, 30288–30304. [Google Scholar] [CrossRef] [PubMed]








| A. Characteristics of Lung Cancer Cases | ||
| Total number | 30 | |
| Median age (range) | 71 (50–88) | |
| Sex | n | (%) |
| Male | 29 | (96.7) |
| Female | 1 | (3.3) |
| Pathological stage | ||
| IA | 5 | (16.7) |
| IB | 9 | (30.0) |
| IIA | 2 | (6.7) |
| IIB | 6 | (20.0) |
| IIIA | 7 | (23.3) |
| IIIB | 1 | (3.3) |
| B. Characteristics of noncancerous tissues | ||
| Total number | 20 | |
| Median age (range) | 70.5 (50–88) | |
| Sex | n | |
| Male | 20 | |
| Female | 0 | |
| Gene ID | Gene Symbol | Description | miR-451a Transfectant Log2 Ratio | miR-451a Target Site | GSE19188 Log FC | TCGA Database5-y OS p-Value | |
|---|---|---|---|---|---|---|---|
| Conserved Site | Poorly Conserved Site | ||||||
| 3796 | KIF2A | kinesin family member 2A | −0.717 | 0 | 1 | 1.01 | 0.043 |
| 23200 | ATP11B | ATPase phospholipid transporting 11B | −0.737 | 0 | 1 | 1.38 | 0.081 |
| 83990 | BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | −0.722 | 0 | 1 | 2.05 | 0.123 |
| 25769 | SLC24A2 | solute carrier family 24 member 2 | −0.734 | 0 | 1 | 1.01 | 0.147 |
| 57405 | SPC25 | SPC25, NDC80 kinetochore complex component | −1.500 | 0 | 1 | 2.42 | 0.263 |
| 4282 | MIF | macrophage migration inhibitory factor | −1.144 | 1 | 0 | 1.58 | 0.357 |
| 84951 | TNS4 | tensin 4 | −0.592 | 0 | 1 | 2.56 | 0.494 |
| 1362 | CPD | carboxypeptidase D | −1.233 | 0 | 1 | 1.07 | 0.598 |
| 23516 | SLC39A14 | solute carrier family 39 member 14 | −1.190 | 0 | 1 | 1.01 | 0.607 |
| 5933 | RBL1 | RB transcriptional corepressor like 1 | −0.922 | 0 | 1 | 1.08 | 0.720 |
| 81796 | SLCO5A1 | solute carrier organic anion transporter family member 5A1 | −0.795 | 0 | 1 | 1.23 | 0.732 |
| 9699 | RIMS2 | regulating synaptic membrane exocytosis 2 | −0.515 | 0 | 1 | 1.98 | 0.815 |
| 64067 | NPAS3 | neuronal PAS domain protein 3 | −1.005 | 0 | 1 | 1.23 | 0.833 |
| 2668 | GDNF | glial cell derived neurotrophic factor | −1.289 | 0 | 1 | 1.01 | 0.866 |
| 23657 | SLC7A11 | solute carrier family 7 member 11 | −0.594 | 0 | 1 | 2.01 | 0.878 |
| A. Immunohistochemical status and characteristics of LUSQ cases | |||||||
| Patient No. | Grade | T | N | M | Pathological Stage | Immunohistochemical Staining Intensity | Immunohistochemical Staining Extensity |
| 23 | 2 | 2 | 1 | 0 | IIB | (+) | (+++) |
| 24 | 2 | 2 | 0 | 0 | IB | (+++) | (+++) |
| 25 | 2 | 1 | 0 | 0 | IA | (+++) | (+++) |
| 26 | 1 | 2 | 1 | 0 | IIB | (++) | (+++) |
| 27 | 2 | 1 | 0 | 0 | IA | (++) | (+++) |
| 28 | 1 | 3 | 0 | 0 | IIB | (++) | (+++) |
| 29 | 1 | 2 | 0 | 0 | IB | (+++) | (+++) |
| 30 | 2 | 2 | 0 | 0 | IB | (+++) | (+++) |
| 31 | 3 | 2 | 0 | 0 | IB | (++) | (+++) |
| 32 | 3 | 2 | 1 | 0 | IIB | (+) | (+++) |
| 33 | 3 | 2 | 0 | 0 | IB | (++) | (+++) |
| 34 | 3 | 2 | 1 | 0 | IIB | (++) | (+++) |
| 35 | 2 | 3 | 1 | 0 | IIIA | (++) | (+++) |
| 36 | 3 | 2 | 1 | 0 | IIA | (++) | (+++) |
| 37 | 3 | 3 | 0 | 0 | IIB | (++) | (+++) |
| 38 | 3 | 2 | 0 | 0 | IB | (+++) | (+++) |
| 39 | 3 | 2 | 1 | 0 | IIB | (+++) | (+++) |
| 40 | 3 | 2 | 0 | 0 | IB | (+++) | (+++) |
| 41 | 2-3 | 3 | 0 | 0 | IIB | (++) | (+++) |
| 42 | 3 | 1 | 2 | 0 | IIIA | (+) | (+++) |
| 43 | 3 | 2 | 0 | 0 | IB | (++) | (+++) |
| 44 | 3 | 2 | 0 | 0 | IB | (++) | (+++) |
| B. Immunohistochemical status of noncancerous cases | |||||||
| Patient No. | Immunohistochemical Staining Intensity | Immunohistochemical Staining Extensity | |||||
| 69 | (++) | (++) | |||||
| 70 | (++) | (+++) | |||||
| 71 | (+) | (++) | |||||
| 72 | (+) | (++) | |||||
| 73 | (+) | (++) | |||||
| 74 | (++) | (+++) | |||||
| 75 | (+) | (++) | |||||
| 76 | (++) | (++) | |||||
| 77 | (+) | (++) | |||||
| 78 | (+) | (+) | |||||
| 79 | (++) | (++) | |||||
| 80 | (++) | (+++) | |||||
| No. of Genes | p-Value | Annotations |
|---|---|---|
| 10 | 5.59 × 10−12 | (KEGG) 04110: Cell cycle |
| 3 | 5.71 × 10−5 | (KEGG) 04115: p53 signaling pathway |
| - | - | (KEGG) 04110: Cell cycle |
| 4 | 7.56 × 10−5 | (KEGG) 04115: p53 signaling pathway |
| 3 | 1.36 × 10−4 | (KEGG) 03030: DNA replication |
| 5 | 1.68 × 10−3 | (KEGG) 05200: Pathways in cancer |
| Gene Symbol | Description | si-KIF2A Transfectant Log2 Ratio | GSE19188 Log FC |
|---|---|---|---|
| Cell cycle | |||
| CCNE2 | cyclin E2 | −0.730 | 2.04 |
| MCM4 | minichromosome maintenance complex component 4 | −0.925 | 3.13 |
| CCNA2 | cyclin A2 | −0.563 | 3.25 |
| CCNE1 | cyclin E1 | −1.099 | 2.23 |
| CHEK1 | checkpoint kinase 1 | −0.586 | 2.76 |
| ORC6 | origin recognition complex subunit 6 | −0.565 | 2.26 |
| CDC6 | cell division cycle 6 | −0.529 | 3.31 |
| ORC1 | origin recognition complex subunit 1 | −0.600 | 2.72 |
| CDC45 | cell division cycle 45 | −0.524 | 3.83 |
| MCM2 | minichromosome maintenance complex component 2 | −0.703 | 2.41 |
| p53 signaling pathway, cell cycle | |||
| CCNE2 | cyclin E2 | −0.730 | 2.04 |
| CCNE1 | cyclin E1 | −1.099 | 2.23 |
| CHECK1 | checkpoint kinase 1 | −0.586 | 2.76 |
| p53 signaling pathway | |||
| CCNE2 | cyclin E2 | −0.730 | 2.04 |
| CCNE1 | cyclin E1 | −1.099 | 2.23 |
| CHEK1 | checkpoint kinase 1 | −0.586 | 2.76 |
| RRM2 | ribonucleotide reductase regulatory subunit M2 | −0.578 | 3.00 |
| DNA replication | |||
| MCM4 | minichromosome maintenance complex component 4 | −0.925 | 3.13 |
| MCM2 | minichromosome maintenance complex component 2 | −0.703 | 2.41 |
| RFC4 | replication factor C subunit 4 | −0.688 | 2.00 |
| Pathway in cancer | |||
| MMP9 | matrix metallopeptidase 9 | −0.531 | 2.04 |
| CCNE2 | cyclin E2 | −0.730 | 2.04 |
| CBLC | Cbl proto-oncogene C | −0.628 | 3.04 |
| CCNE1 | cyclin E1 | −1.099 | 2.23 |
| RAD51 | RAD51 recombinase | −0.594 | 2.09 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uchida, A.; Seki, N.; Mizuno, K.; Yamada, Y.; Misono, S.; Sanada, H.; Kikkawa, N.; Kumamoto, T.; Suetsugu, T.; Inoue, H. Regulation of KIF2A by Antitumor miR-451a Inhibits Cancer Cell Aggressiveness Features in Lung Squamous Cell Carcinoma. Cancers 2019, 11, 258. https://doi.org/10.3390/cancers11020258
Uchida A, Seki N, Mizuno K, Yamada Y, Misono S, Sanada H, Kikkawa N, Kumamoto T, Suetsugu T, Inoue H. Regulation of KIF2A by Antitumor miR-451a Inhibits Cancer Cell Aggressiveness Features in Lung Squamous Cell Carcinoma. Cancers. 2019; 11(2):258. https://doi.org/10.3390/cancers11020258
Chicago/Turabian StyleUchida, Akifumi, Naohiko Seki, Keiko Mizuno, Yasutaka Yamada, Shunsuke Misono, Hiroki Sanada, Naoko Kikkawa, Tomohiro Kumamoto, Takayuki Suetsugu, and Hiromasa Inoue. 2019. "Regulation of KIF2A by Antitumor miR-451a Inhibits Cancer Cell Aggressiveness Features in Lung Squamous Cell Carcinoma" Cancers 11, no. 2: 258. https://doi.org/10.3390/cancers11020258





