Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis
Abstract
:1. Introduction
2. Results
2.1. Connexin mRNA Expression in Human Lung Tumours and Normal Healthy Tissue
2.2. Regulation of Connexins at the DNA and mRNA Levels in Relation to Lung Cancer
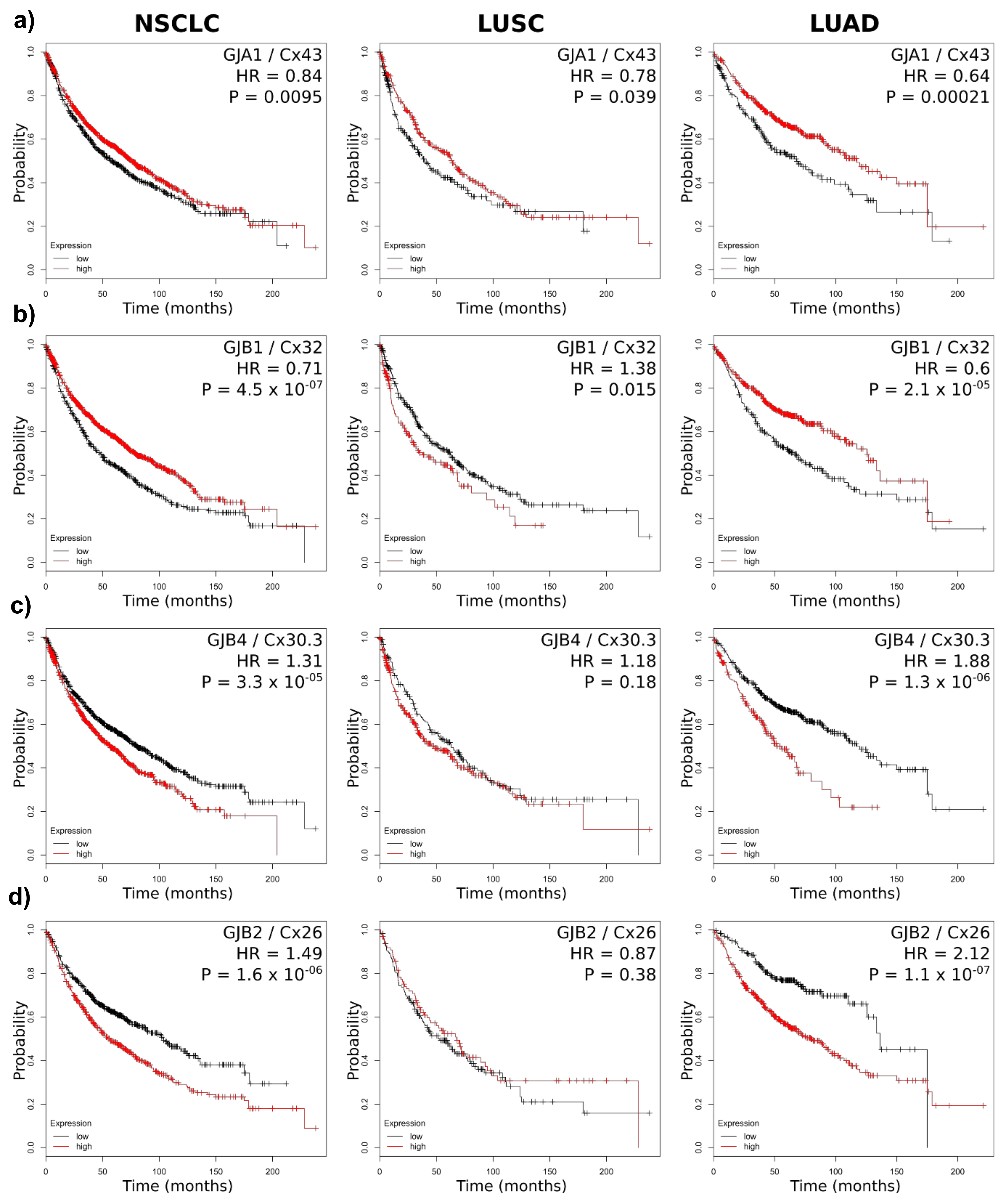
2.3. Connexin mRNA Expression Is Associated with Both Poor and Good Prognosis, Which Depends on Both Lung Cancer Subtype and Connexin Isoform
2.4. Connexin Protein Expression and Subcellular Localization
3. Discussion
4. Materials and Methods
4.1. Immunohistochemistry
4.2. Statistics
4.3. Human Tissue Specimens and Tissue Microarray
4.4. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Johnstone, S.; Vidal-Brime, L.; Lynn, K.S.; Koval, M. Connexins: Synthesis, post-translational modifications, and trafficking in health and disease. Int. J. Mol. Sci. 2018, 19, 1296. [Google Scholar] [CrossRef] [PubMed]
- Schalper, K.A.; Carvajal-Hausdorf, D.; Oyarzo, M.P. Possible role of hemichannels in cancer. Front. Physiol. 2014, 5, 237. [Google Scholar] [CrossRef] [PubMed]
- Leithe, E.; Mesnil, M.; Aasen, T. The connexin 43 c-terminus: A tail of many tales. Biochim. Biophys. Acta 2018, 1860, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T. Connexins: Junctional and non-junctional modulators of proliferation. Cell Tissue Res. 2015, 360, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Smyth, J.W.; Shaw, R.M. Autoregulation of connexin43 gap junction formation by internally translated isoforms. Cell Rep. 2013, 5, 611–618. [Google Scholar] [CrossRef]
- Salat-Canela, C.; Sese, M.; Peula, C.; Ramon y Cajal, S.; Aasen, T. Internal translation of the connexin 43 transcript. Cell Commun. Signal. 2014, 12, 31. [Google Scholar] [CrossRef]
- Ul-Hussain, M.; Olk, S.; Schoenebeck, B.; Wasielewski, B.; Meier, C.; Prochnow, N.; May, C.; Galozzi, S.; Marcus, K.; Zoidl, G.; et al. Internal ribosomal entry site (ires) activity generates endogenous carboxyl-terminal domains of cx43 and is responsive to hypoxic conditions. J. Biol. Chem. 2014, 289, 20979–20990. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, S.S.; Xiao, S.; Basheer, W.A.; Baum, R.; Epifantseva, I.; Hong, T.; Shaw, R.M. Cx43 isoform gja1-20k promotes microtubule dependent mitochondrial transport. Front. Physiol. 2017, 8, 905. [Google Scholar] [CrossRef]
- Basheer, W.A.; Xiao, S.; Epifantseva, I.; Fu, Y.; Kleber, A.G.; Hong, T.; Shaw, R.M. Gja1-20k arranges actin to guide cx43 delivery to cardiac intercalated discs. Circ. Res. 2017, 121, 1069–1080. [Google Scholar] [CrossRef]
- Basheer, W.; Shaw, R. The “tail” of connexin43: An unexpected journey from alternative translation to trafficking. Biochim. Biophys. Acta 2016, 1863, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- James, C.C.; Zeitz, M.J.; Calhoun, P.J.; Lamouille, S.; Smyth, J.W. Altered translation initiation of gja1 limits gap junction formation during epithelial-mesenchymal transition. Mol. Biol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Kotini, M.; Barriga, E.H.; Leslie, J.; Gentzel, M.; Rauschenberger, V.; Schambon, A.; Mayor, R. Gap junction protein connexin-43 is a direct transcriptional regulator of n-cadherin in vivo. Nat. Commun. 2018, 9, 3846. [Google Scholar] [CrossRef] [PubMed]
- Losa, D.; Chanson, M. The lung communication network. Cell. Mol. Life Sci. 2015, 72, 2793–2808. [Google Scholar] [CrossRef] [PubMed]
- Losa, D.; Chanson, M.; Crespin, S. Connexins as therapeutic targets in lung disease. Expert Opin. Ther. Targets 2011, 15, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Mesnil, M.; Naus, C.C.; Lampe, P.D.; Laird, D.W. Gap junctions and cancer: Communicating for 50 years. Nat. Rev. Cancer 2016, 16, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.P.; Wei, R. Effects of cx43 gene modification on the proliferation and migration of the human lung squamous carcinoma cell line nci-h226. Genet. Mol. Res. 2015, 14, 13110–13119. [Google Scholar] [CrossRef]
- Xu, H.T.; Li, Q.C.; Zhang, Y.X.; Zhao, Y.; Liu, Y.; Yang, Z.Q.; Wang, E.H. Connexin 43 recruits e-cadherin expression and inhibits the malignant behaviour of lung cancer cells. Folia Histochem. Cytobiol. 2008, 46, 315–321. [Google Scholar] [CrossRef]
- Zhao, W.; Han, H.B.; Zhang, Z.Q. Suppression of lung cancer cell invasion and metastasis by connexin43 involves the secretion of follistatin-like 1 mediated via histone acetylation. Int. J. Biochem. Cell Biol. 2011, 43, 1459–1468. [Google Scholar] [CrossRef]
- Ruch, R.J. Connexin43 suppresses lung cancer stem cells. Cancers 2019, 11, 175. [Google Scholar] [CrossRef]
- Chen, Y.; Huhn, D.; Knosel, T.; Pacyna-Gengelbach, M.; Deutschmann, N.; Petersen, I. Downregulation of connexin 26 in human lung cancer is related to promoter methylation. Int. J. Cancer 2005, 113, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, C.; Li, Y.; Fu, X.; Xie, Y.; Li, Y.; Huang, Y. Cx31.1 acts as a tumour suppressor in non-small cell lung cancer (nsclc) cell lines through inhibition of cell proliferation and metastasis. J. Cell. Mol. Med. 2012, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ruan, Z.; Yang, X.; Chu, K.; Wu, H.; Li, Y.; Huang, Y. Connexin 31.1 degradation requires the clathrin-mediated autophagy in nsclc cell h1299. J. Cell. Mol. Med. 2015, 19, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Fukumoto, K.; Hada, S.; Hagiwara, H.; Fujimoto, E.; Negishi, E.; Ueno, K.; Yano, T. Enhancing effect of connexin 32 gene on vinorelbine-induced cytotoxicity in a549 lung adenocarcinoma cells. Cancer Chemother. Pharm. 2007, 60, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Hada, S.; Sato, H.; Virgona, N.; Hagiwara, H.; Saito, T.; Suzuki, K.; Asano, R.; Yano, T. Connexin 32 expression reduces malignant phenotype in human a549 adenocarcinoma cells: Implication of src involvement. Oncol. Rep. 2006, 16, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- King, T.J.; Lampe, P.D. The gap junction protein connexin32 is a mouse lung tumor suppressor. Cancer Res. 2004, 64, 7191–7196. [Google Scholar] [CrossRef] [PubMed]
- King, T.J.; Lampe, P.D. Mice deficient for the gap junction protein connexin32 exhibit increased radiation-induced tumorigenesis associated with elevated mitogen-activated protein kinase (p44/erk1, p42/erk2) activation. Carcinogenesis 2004, 25, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.I.; Hirabayashi, Y.; Kawasaki, Y.; Tsuboi, I.; Ott, T.; Kodama, Y.; Kanno, J.; Kim, D.Y.; Willecke, K.; Inoue, T. Exacerbation of benzene pneumotoxicity in connexin 32 knockout mice: Enhanced proliferation of cyp2e1-immunoreactive alveolar epithelial cells. Toxicology 2004, 195, 19–29. [Google Scholar] [CrossRef]
- Avanzo, J.L.; Mennecier, G.; Mesnil, M.; Hernandez-Blazquez, F.J.; Fukumasu, H.; da Silva, T.C.; Rao, K.V.; Dagli, M.L. Deletion of a single allele of cx43 is associated with a reduction in the gap junctional intercellular communication and increased cell proliferation of mouse lung pneumocytes type ii. Cell Prolif. 2007, 40, 411–421. [Google Scholar] [CrossRef]
- De Oliveira, K.D.; Tedardi, M.V.; Cogliati, B.; Dagli, M.L. Higher incidence of lung adenocarcinomas induced by dmba in connexin 43 heterozygous knockout mice. BioMed Res. Int. 2013, 2013, 618475. [Google Scholar] [CrossRef]
- Fukumasu, H.; Avanzo, J.L.; Sanches, D.S.; Mennecier, G.; Mori, C.M.; Dagli, M.L. Higher susceptibility of spontaneous and nnk-induced lung neoplasms in connexin 43 deficient cd1 x aj f1 mice: Paradoxical expression of connexin 43 during lung carcinogenesis. Mol. Carcinog. 2013, 52, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Vinken, M. Regulation of connexin signaling by the epigenetic machinery. Biochim. Biophys. Acta 2015, 1859, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Salat-Canela, C.; Munoz, M.J.; Sese, M.; Ramon y Cajal, S.; Aasen, T. Post-transcriptional regulation of connexins. Biochem. Soc. Trans. 2015, 43, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.T.; Cheng, Y.W.; Chou, M.C.; Sen-Lin, T.; Lai, W.W.; Ho, W.L.; Lee, H. The correlation between aberrant connexin 43 mrna expression induced by promoter methylation and nodal micrometastasis in non-small cell lung cancer. Clin. Cancer Res. 2003, 9, 4200–4204. [Google Scholar] [PubMed]
- Jinn, Y.; Inase, N. Connexin 43, e-cadherin, β-catenin and zo-1 expression, and aberrant methylation of the connexin 43 gene in nsclc. Anticancer Res. 2010, 30, 2271–2278. [Google Scholar] [PubMed]
- Zhang, W.; Li, H.G.; Fan, M.J.; Lv, Z.Q.; Shen, X.M.; He, X.X. Expressions of connexin 32 and 26 and their correlation to prognosis of non-small cell lung cancer. Ai Zheng 2009, 28, 173–176. [Google Scholar] [PubMed]
- Du, G.; Yang, Y.; Zhang, Y.; Sun, T.; Liu, W.; Wang, Y.; Li, J.; Zhang, H. Thrombocytosis and immunohistochemical expression of connexin 43 at diagnosis predict survival in advanced non-small-cell lung cancer treated with cisplatin-based chemotherapy. Cancer Chemother. Pharm. 2013, 71, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Mesnil, M.; Aasen, T.; Boucher, J.; Chepied, A.; Cronier, L.; Defamie, N.; Kameritsch, P.; Laird, D.W.; Lampe, P.D.; Lathia, J.D.; et al. An update on minding the gap in cancer. Biochim. Biophys. Acta Biomembr. 2018, 1860, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Naus, C.C.; Laird, D.W. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer 2010, 10, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, C.; Li, L.; Dong, S.; Zhang, N.; Tong, X. Cx43 reverses the resistance of a549 lung adenocarcinoma cells to cisplatin by inhibiting emt. Oncol. Rep. 2014, 31, 2751–2758. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qin, G.; Luo, M.; Chen, J.; Zhang, Q.; Li, L.; Pan, L.; Qin, S. Reciprocal positive regulation between cx26 and pi3k/akt pathway confers acquired gefitinib resistance in nsclc cells via gjic-independent induction of emt. Cell Death Dis. 2015, 6, e1829. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Koma, Y.; Uchino, K.; Okada, T.; Ohbayashi, C.; Tsubota, N.; Okada, M. Increased expression of connexin 26 in the invasive component of lung squamous cell carcinoma: Significant correlation with poor prognosis. Cancer Lett. 2006, 234, 239–248. [Google Scholar] [CrossRef] [PubMed]
- el-Sabban, M.E.; Pauli, B.U. Adhesion-mediated gap junctional communication between lung-metastatatic cancer cells and endothelium. Invasion Metastasis 1994, 14, 164–176. [Google Scholar] [PubMed]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cgamp transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Menachem, A.; Makovski, V.; Bodner, O.; Pasmanik-Chor, M.; Stein, R.; Shomron, N.; Kloog, Y. Intercellular transfer of small rnas from astrocytes to lung tumor cells induces resistance to chemotherapy. Oncotarget 2016, 7, 12489–12504. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Wu, J.I.; Tseng, C.W.; Chen, H.J.; Wang, L.H. Gjb4 serves as a novel biomarker for lung cancer and promotes metastasis and chemoresistance via src activation. Oncogene 2019, 38, 822–837. [Google Scholar] [CrossRef] [PubMed]
- Crespin, S.; Fromont, G.; Wager, M.; Levillain, P.; Cronier, L.; Monvoisin, A.; Defamie, N.; Mesnil, M. Expression of a gap junction protein, connexin43, in a large panel of human gliomas: New insights. Cancer Med. 2016, 5, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Sirnes, S.; Bruun, J.; Kolberg, M.; Kjenseth, A.; Lind, G.E.; Svindland, A.; Brech, A.; Nesbakken, A.; Lothe, R.A.; Leithe, E.; et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int. J. Cancer 2012, 131, 570–581. [Google Scholar] [CrossRef]
- Cesen-Cummings, K.; Fernstrom, M.J.; Malkinson, A.M.; Ruch, R.J. Frequent reduction of gap junctional intercellular communication and connexin43 expression in human and mouse lung carcinoma cells. Carcinogenesis 1998, 19, 61–67. [Google Scholar] [CrossRef]
- de Feijter, A.W.; Matesic, D.F.; Ruch, R.J.; Guan, X.; Chang, C.C.; Trosko, J.E. Localization and function of the connexin 43 gap-junction protein in normal and various oncogene-expressing rat liver epithelial cells. Mol. Carcinog. 1996, 16, 203–212. [Google Scholar] [CrossRef]
- Chen, X.; Kong, X.; Zhuang, W.; Teng, B.; Yu, X.; Hua, S.; Wang, S.; Liang, F.; Ma, D.; Zhang, S.; et al. Dynamic changes in protein interaction between akap95 and cx43 during cell cycle progression of a549 cells. Sci. Rep. 2016, 6, 21224. [Google Scholar] [CrossRef] [PubMed]
- Mennecier, G.; Derangeon, M.; Coronas, V.; Herve, J.C.; Mesnil, M. Aberrant expression and localization of connexin43 and connexin30 in a rat glioma cell line. Mol. Carcinog. 2008, 47, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Bragelmann, J.; Kryukov, I.; Saraiva-Agostinho, N.; Perner, S. Firebrowser: An r client to the broad institute’s firehose pipeline. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Men, C.; Chai, H.; Song, X.; Li, Y.; Du, H.; Ren, Q. Identification of DNA methylation associated gene signatures in endometrial cancer via integrated analysis of DNA methylation and gene expression systematically. J. Gynecol. Oncol. 2017, 28, e83. [Google Scholar] [CrossRef] [PubMed]
- Shinawi, T.; Hill, V.K.; Krex, D.; Schackert, G.; Gentle, D.; Morris, M.R.; Wei, W.; Cruickshank, G.; Maher, E.R.; Latif, F. DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics 2013, 8, 149–156. [Google Scholar] [CrossRef]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Tittarelli, A.; Guerrero, I.; Tempio, F.; Gleisner, M.A.; Avalos, I.; Sabanegh, S.; Ortiz, C.; Michea, L.; Lopez, M.N.; Mendoza-Naranjo, A.; et al. Overexpression of connexin 43 reduces melanoma proliferative and metastatic capacity. Br. J. Cancer 2015, 113, 259–267. [Google Scholar] [CrossRef]
- Poyet, C.; Buser, L.; Roudnicky, F.; Detmar, M.; Hermanns, T.; Mannhard, D.; Hohn, A.; Ruschoff, J.; Zhong, Q.; Sulser, T.; et al. Connexin 43 expression predicts poor progression-free survival in patients with non-muscle invasive urothelial bladder cancer. J. Clin. Pathol. 2015, 68, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Moarii, M.; Boeva, V.; Vert, J.P.; Reyal, F. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genom. 2015, 16, 873. [Google Scholar] [CrossRef] [PubMed]
- King, T.J.; Gurley, K.E.; Prunty, J.; Shin, J.L.; Kemp, C.J.; Lampe, P.D. Deficiency in the gap junction protein connexin32 alters p27kip1 tumor suppression and mapk activation in a tissue-specific manner. Oncogene 2005, 24, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Luo, Y.; Mao, N.; Huang, G.; Teng, C.; Wang, H.; Wu, J.; Liao, X.; Yang, J. Cancer-associated fibroblasts accelerate malignant progression of non-small cell lung cancer via connexin 43-formed unidirectional gap junctional intercellular communication. Cell. Physiol. Biochem. 2018, 51, 315–336. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Bhattacharya, S.; Kalyan, G.; Hazra, S. Cadherin profiling for therapeutic interventions in epithelial mesenchymal transition (emt) and tumorigenesis. Exp. Cell Res. 2018, 368, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Zhang, S.; Dong, X.; Tian, D.; Cui, Z.; Qiu, X. Prognostic significance of twist and n-cadherin expression in nsclc. PLoS ONE 2013, 8, e62171. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Yoshino, I.; Yamaguchi, R.; Shimamura, T.; Nagasaki, M.; Imoto, S.; Niida, A.; Koizumi, F.; Kohno, T.; Yokota, J.; et al. N-cadherin expression is a potential survival mechanism of gefitinib-resistant lung cancer cells. Am. J. Cancer Res. 2011, 1, 823–833. [Google Scholar]


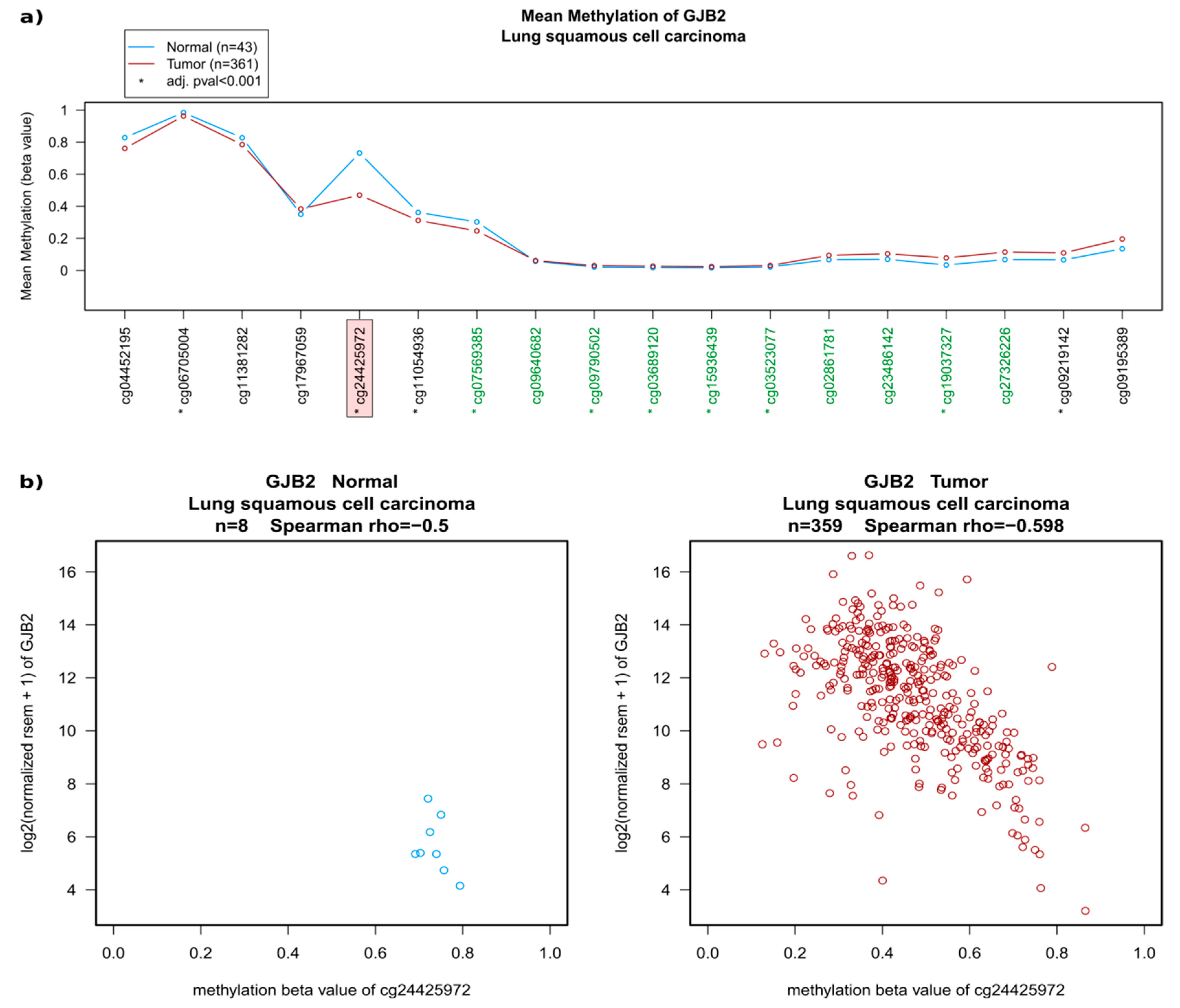


| Regulation | Cx | Gene | LUAD | LUSC |
|---|---|---|---|---|
| Up | Cx46 | GJA3 | 1.74 | 35.6 |
| Cx59 | GJA9 | 3.01 | 1.82 | |
| Cx26 | GJB2 | 14.4 | 63.5 | |
| Cx31 | GJB3 | 3.27 | 34.4 | |
| Cx30.3 | GJB4 | 1.88 | 29.4 | |
| Cx31.1 | GJB5 | 1.66 | 59.9 | |
| Cx30 | GJB6 | 7.47 | 289 | |
| Down | Cx37 | GJA4 | 0.36 | 0.18 |
| Cx40 | GJA5 | 0.32 | 0.17 | |
| Cx50 | GJA8 | 0.83 | 0.97 | |
| Cx45 | GJC1 | 0.67 | 0.76 | |
| Cx47 | GJC2 | 0.325 | 0.234 | |
| Cx30.2 | GJC3 | 0.817 | 0.61 | |
| Cx31.9 | GJD3 | 0.52 | 0.43 | |
| Cx40.1 | GJD4 | 0.89 | 0.96 | |
| Mixed | Cx43 | GJA1 | 0.31 | 1.33 |
| Cx32 | GJB1 | 1.49 | 0.064 | |
| Cx25 | GJB7 | 0.41 | 7.18 | |
| Cx36 | GJD2 | 0.56 | 2.9 |
| Gene | Connexin | HR NSCLC | OS Change (Months) High Cx | HR LUSC | OS Change (Months) High Cx | HR LUAD | OS Change (Months) High Cx |
|---|---|---|---|---|---|---|---|
| GJA1 | Cx43 | 0.84 * | +19 | 0.78 | +24 | 0.64 * | +49 |
| GJA3 | Cx46 | 1.42 * | −41 | 0.77 | +27 | 1.48 * | −42 |
| GJA4 | Cx37 | 0.67 * | +43 | 0.83 | +22 | 0.67 * | +45 |
| GJA5 | Cx40 | 1.30 * | −23 | 1.20 | −4 | 1.86 * | −69 |
| GJA8 | Cx50 | 1.18 | −11 | 1.16 | −17 | 0.60 * | +59 |
| GJA10 | Cx62 | 1.20 | −28 | 0.77 | +24 | 1.62 * | −52 |
| GJB1 | Cx32 | 0.71 * | +31 | 1.38 | −24 | 0.60 * | +59 |
| GJB3 | Cx31 | 1.56 * | −37 | 1.27 | −21 | 2.39 * | −114 |
| GJB4 | Cx30.3 | 1.31 * | −23 | 1.18 | −15 | 1.88 * | −62 |
| GJB5 | Cx31.1 | 1.52 * | −36 | 1.12 | −12 | 1.63 * | −57 |
| GJB6 | Cx30 | 1.40 * | −38 | 0.75 | +29 | 0.82 | +11 |
| GJB2 | Cx26 | 1.49 * | −47 | 0.87 | +16 | 2.12 * | −56 |
| GJC1 | Cx45 | 0.73 * | +36 | 1.22 | −15 | 0.66 * | +37 |
| GJC2 | Cx47 | 1.13 | −6 | 0.81 | +27 | 1.45 * | −41 |
| GJC3 | Cx30.2 | 1.26 * | −27 | 0.78 | +8 | 1.76 * | −53 |
| GJD2 | Cx36 | 1.25 * | −18 | 1.14 | −11 | 2.13 * | −68 |
| GJD3 | Cx31.9 | 0.68 * | +39 | 0.66 * | +24 | 1.26 | −44 |
| GJD4 | Cx40.1 | 1.25 * | −22 | 0.83 | +27 | 1.49 * | −33 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aasen, T.; Sansano, I.; Montero, M.Á.; Romagosa, C.; Temprana-Salvador, J.; Martínez-Marti, A.; Moliné, T.; Hernández-Losa, J.; Ramón y Cajal, S. Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis. Cancers 2019, 11, 320. https://doi.org/10.3390/cancers11030320
Aasen T, Sansano I, Montero MÁ, Romagosa C, Temprana-Salvador J, Martínez-Marti A, Moliné T, Hernández-Losa J, Ramón y Cajal S. Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis. Cancers. 2019; 11(3):320. https://doi.org/10.3390/cancers11030320
Chicago/Turabian StyleAasen, Trond, Irene Sansano, Maria Ángeles Montero, Cleofé Romagosa, Jordi Temprana-Salvador, Alexandre Martínez-Marti, Teresa Moliné, Javier Hernández-Losa, and Santiago Ramón y Cajal. 2019. "Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis" Cancers 11, no. 3: 320. https://doi.org/10.3390/cancers11030320
APA StyleAasen, T., Sansano, I., Montero, M. Á., Romagosa, C., Temprana-Salvador, J., Martínez-Marti, A., Moliné, T., Hernández-Losa, J., & Ramón y Cajal, S. (2019). Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis. Cancers, 11(3), 320. https://doi.org/10.3390/cancers11030320







