Comparative Cytogenetic Abnormalities in Paired Choroidal Melanoma Samples Obtained Before and After Proton Beam Irradiation by Transscleral Fine-Needle Aspiration Biopsy and Endoresection
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics
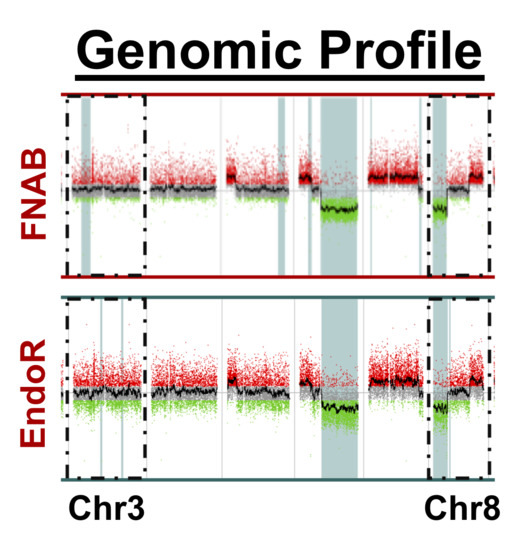
2.2. Cytogenetic Analyses
3. Discussion
- The material retrieved during endoresection has a significantly higher yield, in term of contributive samples, than the biopsies retrieved by fine-needle aspiration;
- There was a high concordance rate between paired samples, especially regarding chromosomes associated with prognosis (i.e., chromosomes 3 and 8);
- The discordance observed in a minority of samples may be related to tumor cell content of sample heterogeneity;
- Cytogenetic alterations are not affected by irradiation, at least within the 2–3 month delay between irradiation and endoresection.
4. Materials and Methods
4.1. Patients
- Conservative management by proton beam irradiation;
- Fine-needle aspiration biopsy for cytogenetic analysis performed at the time of tantalum clip surgery before irradiation;
- Endoresection of the tumor scar performed within three months of irradiation, with cytogenetic analysis of the endoresection material.
4.2. Surgical Procedures
4.2.1. Transscleral Fine-Needle Aspiration Biopsy
4.2.2. Endoresection
4.3. Irradiation Technique
4.4. Cytogenetic Analyses
4.4.1. DNA Extraction
4.4.2. DNA Quantity and Quality Assessment
4.4.3. Array Comparative Genomic Hybridization (CGH)
4.4.4. Next-Generation Sequencing (NGS) Panel
4.4.5. Chromosome Abnormalities
- -
- Presence of total or partial chromosome 3 loss and/or chromosome 8q gain;
- -
- Absence of alteration on chromosomes 3 and 8, but presence of other chromosomal alterations or identification of GNAQ and/or GNA11 mutations on the NGS Panel, confirming the diagnosis of choroidal melanoma;
- -
- Non-contributive sample: insufficient DNA amount or quality.
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Damato, B.; Eleuteri, A.; Taktak, A.F.G.; Coupland, S.E. Estimating prognosis for survival after treatment of choroidal melanoma. Prog. Retin. Eye Res. 2011, 30, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Dogrusöz, M.; Jager, M.J.; Damato, B. Uveal Melanoma Treatment and Prognostication. Asia-Pac. J. Ophthalmol. 2017, 6, 186–196. [Google Scholar]
- Croce, M.; Ferrini, S.; Pfeffer, U.; Gangemi, R. Targeted Therapy of Uveal Melanoma: Recent Failures and New Perspectives. Cancers 2019, 11, 846. [Google Scholar] [CrossRef] [PubMed]
- Castet, F.; Garcia-Mulero, S.; Sanz-Pamplona, R.; Cuellar, A.; Casanovas, O.; Caminal, J.; Piulats, J. Uveal Melanoma, Angiogenesis and Immunotherapy, Is There Any Hope? Cancers 2019, 11, 834. [Google Scholar] [CrossRef] [PubMed]
- Violanti, S.S.; Bononi, I.; Gallenga, C.E.; Martini, F.; Tognon, M.; Perri, P. New Insights into Molecular Oncogenesis and Therapy of Uveal Melanoma. Cancers 2019, 11, 694. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Royer-Bertrand, B.; Torsello, M.; Rimoldi, D.; El Zaoui, I.; Cisarova, K.; Pescini-Gobert, R.; Raynaud, F.; Zografos, L.; Schalenbourg, A.; Speiser, D.; et al. Comprehensive Genetic Landscape of Uveal Melanoma by Whole-Genome Sequencing. Am. J. Hum. Genet. 2016, 99, 1190–1198. [Google Scholar] [CrossRef]
- Amirouchene-Angelozzi, N.; Schoumacher, M.; Stern, M.-H.; Cassoux, N.; Desjardins, L.; Piperno-Neumann, S.; Lantz, O.; Roman-Roman, S. Upcoming translational challenges for uveal melanoma. Br. J. Cancer 2015, 113, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Naus, N.C.; Verhoeven, A.C.A.; Van Drunen, E.; Slater, R.; Mooy, C.M.; Paridaens, D.A.; Luyten, G.P.M.; De Klein, A. Detection of genetic prognostic markers in uveal melanoma biopsies using fluorescence in situ hybridization. Clin. Cancer Res. 2002, 8, 534–539. [Google Scholar]
- Sandinha, M.T.; Farquharson, M.A.; Roberts, F. Identification of monosomy 3 in choroidal melanoma by chromosome in situ hybridisation. Br. J. Ophthalmol. 2004, 88, 1527–1532. [Google Scholar] [CrossRef]
- Damato, B.; Dopierala, J.A.; Coupland, S.E. Genotypic Profiling of 452 Choroidal Melanomas with Multiplex Ligation-Dependent Probe Amplification. Clin. Cancer Res. 2010, 16, 6083–6092. [Google Scholar] [CrossRef]
- Cassoux, N.; Rodrigues, M.J.; Plancher, C.; Asselain, B.; Levy-Gabriel, C.; Lumbroso-Le Rouic, L.; Piperno-Neumann, S.; Dendale, R.; Sastre, X.; Desjardins, L.; et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br. J. Ophthalmol. 2014, 98, 769–774. [Google Scholar] [CrossRef]
- Shields, C.L.; Ganguly, A.; Bianciotto, C.G.; Turaka, K.; Tavallali, A.; Shields, J.A. Prognosis of Uveal Melanoma in 500 Cases Using Genetic Testing of Fine-Needle Aspiration Biopsy Specimens. Ophthalmology 2011, 118, 396–401. [Google Scholar] [CrossRef]
- Hussain, R.N.; Kalirai, H.; Groenewald, C.; Kacperek, A.; Errington, R.D.; Coupland, S.E.; Heimann, H.; Damato, B. Prognostic Biopsy of Choroidal Melanoma after Proton Beam Radiation Therapy. Ophthalmology 2016, 123, 2264–2265. [Google Scholar] [CrossRef]
- Shields, C.L.; Say, E.A.T.; Hasanreisoglu, M.; Saktanasate, J.; Lawson, B.M.; Landy, J.E.; Badami, A.U.; Sivalingam, M.D.; Mashayekhi, A.; Shields, J.A.; et al. Cytogenetic Abnormalities in Uveal Melanoma Based on Tumor Features and Size in 1059 Patients. Ophthalmology 2017, 124, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Anthony Say, E.T.; Hasanreisoglu, M.; Saktanasate, J.; Lawson, B.M.; Landy, J.E.; Badami, A.U.; Sivalingam, M.D.; Hauschild, A.J.; House, R.J.; et al. Personalized Prognosis of Uveal Melanoma Based on Cytogenetic Profile in 1059 Patients over an 8-Year Period. Ophthalmology 2017, 124, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Seider, M.I.; Mruthyunjaya, P. Molecular prognostics for uveal melanoma. Retina 2018, 38, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Sellam, A.; Desjardins, L.; Barnhill, R.; Plancher, C.; Asselain, B.; Savignoni, A.; Pierron, G.; Cassoux, N. Fine Needle Aspiration Biopsy in Uveal Melanoma: Technique, Complications, and Outcomes. Am. J. Ophthalmol. 2016, 162, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Angi, M.; Kalirai, H.; Taktak, A.; Hussain, R.; Groenewald, C.; Damato, B.E.; Heimann, H.; Coupland, S.E. Prognostic biopsy of choroidal melanoma: an optimised surgical and laboratory approach. Br. J. Ophthalmol. 2017, 101, 1143–1146. [Google Scholar] [CrossRef]
- Bechrakis, N.E.; Höcht, S.; Martus, P.; Kreusel, K.M.; Heese, J.; Foerster, M.H. Endoresection following proton beam irradiation of large uveal melanomas. Ophthalmologe 2004, 101, 370–376. (In German) [Google Scholar] [CrossRef] [PubMed]
- Cassoux, N.; Cayette, S.; Plancher, C.; Lumbroso-Le Rouic, L.; Levy-Gabriel, C.; Asselain, B.; Sastre, X.; Couturier, J.; Arrufat, S.; Piperno-Neumann, S.; et al. Choroidal Melanoma: Does endoresection prevent neovascular glaucoma in patient treated with proton beam irradiation? Retina 2013, 33, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B. AJCC Cancer Staging Manual; Springer: Berlin, Germany, 2017; ISBN 9783319406176. [Google Scholar]
- McCannel, T.A.; Chang, M.Y.; Burgess, B.L. Multi-Year Follow-up of Fine-Needle Aspiration Biopsy in Choroidal Melanoma. Ophthalmology 2012, 119, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Bagger, M.; Smidt-Nielsen, I.; Andersen, M.K.; Jensen, P.K.; Heegaard, S.; Andersen, K.K.; Friis, S.; Kiilgaard, J.F. Long-Term Metastatic Risk after Biopsy of Posterior Uveal Melanoma. Ophthalmology 2018, 125, 1969–1976. [Google Scholar] [CrossRef] [PubMed]
- Mashayekhi, A.; Lim, R.P.; Shields, C.L.; Eagle, R.C.; Shields, J.A. Extraocular extension of ciliochoroidal melanoma after transscleral fine-needle aspiration biopsy. Retin. Cases Brief Rep. 2016, 10, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Schefler, A.C.; Gologorsky, D.; Marr, B.P.; Shields, C.L.; Zeolite, I.; Abramson, D.H. Extraocular Extension of Uveal Melanoma After Fine-Needle Aspiration, Vitrectomy, and Open Biopsy. JAMA Ophthalmol. 2013, 131, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Coupland, S.E.; Kalirai, H.; Ho, V.; Thornton, S.; Damato, B.E.; Heimann, H. Concordant chromosome 3 Results: In paired choroidal melanoma biopsies and subsequent tumour resection specimens. Br. J. Ophthalmol. 2015, 99, 1444–1450. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Marshall, T.I.; Currell, F.J.; Kacperek, A.; Schettino, G.; Prise, K.M. Variations in the Processing of DNA Double-Strand Breaks Along 60-MeV Therapeutic Proton Beams. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Chen, B.P.; Li, W.; Perko, Z.; Wang, Y.; Testa, M.; Schneider, R.; Lu, H.-M.; Gerweck, L.E. The Relative Biological Effect of Spread-Out Bragg Peak Protons in Sensitive and Resistant Tumor Cells. Int. J. Part. Ther. 2017, 4, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Friedland, W.; Kundrát, P.; Schmitt, E.; Becker, J.; Li, W. Modeling DNA damage by photons and light ions over energy ranges used in medical applications. Radiat. Prot. Dosimetry 2019, 183, 84–88. [Google Scholar] [CrossRef]
- Taleei, R. Modelling DSB repair kinetics for dna damage induced by proton and carbon ions. Radiat. Prot. Dosimetry 2019, 183, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Mobuchon, L.; Houy, A.; Alsafadi, S.; Baulande, S.; Mariani, O.; Marande, B.; Rais, K.A.; Van der Kooij, M.K.; Kapiteijn, E.H.W.; et al. Evolutionary routes in metastatic uveal melanomas depend on MBD4 alterations. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Sandinha, T.; Farquharson, M.; McKay, I.; Roberts, F. Correlation of Heterogeneity for Chromosome 3 Copy Number with Cell Type in Choroidal Melanoma of Mixed-Cell Type. Investig. Opthalmol. Vis. Sci. 2006, 47, 5177–5180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mensink, H.W.; Vaarwater, J.; Kiliç, E.; Naus, N.C.; Mooy, N.; Luyten, G.; Brüggenwirth, H.T.; Paridaens, D.; de Klein, A. Chromosome 3 Intratumor Heterogeneity in Uveal Melanoma. Investig. Opthalmol. Vis. Sci. 2009, 50, 500–504. [Google Scholar] [CrossRef]
- Chang, M.Y.; Rao, N.P.; Burgess, B.L.; Johnson, L.; McCannel, T.A. Heterogeneity of monosomy 3 in fine needle aspiration biopsy of choroidal melanoma. Mol. Vis. 2013, 19, 1892–1900. [Google Scholar]
- Augsburger, B.D.; Augsburger, J.J. Frequency and Implications of Discordant Gene Expression Profile Class in Posterior Uveal Melanomas Sampled by Fine Needle Aspiration Biopsy. Am. J. Ophthalmol. 2015, 159, 248–256. [Google Scholar] [CrossRef]
- Miller, A.K.; Benage, M.J.; Wilson, D.J.; Skalet, A.H. Uveal Melanoma with Histopathologic Intratumoral Heterogeneity Associated with Gene Expression Profile Discordance. Ocul. Oncol. Pathol. 2017, 3, 156–160. [Google Scholar] [CrossRef]
- Herwig-Carl, M.C.; Sharma, A.; Höller, T.; Holz, F.G.; Schlitter, A.M.; Loeffler, K.U. Spatial intratumor heterogeneity in uveal melanoma: Tumor cell subtypes with a presumed invasive potential exhibit a particular epigenetic staining reaction. Exp. Eye Res. 2019, 182, 175–181. [Google Scholar] [CrossRef]
- Höglund, M.; Gisselsson, D.; Hansen, G.B.; White, V.A.; Säll, T.; Mitelman, F.; Horsman, D. Dissecting karyotypic patterns in malignant melanomas: Temporal clustering of losses and gains in melanoma karyotypic evolution. Int. J. Cancer 2004, 108, 57–65. [Google Scholar] [CrossRef]
- Damato, B.; Dopierala, J.; Klaasen, A.; van Dijk, M.; Sibbring, J.; Coupland, S.E. Multiplex Ligation-Dependent Probe Amplification of Uveal Melanoma: Correlation with Metastatic Death. Investig. Opthalmol. Vis. Sci. 2009, 50, 3048–3055. [Google Scholar] [CrossRef]
- Chicard, M.; Boyault, S.; Colmet Daage, L.; Richer, W.; Gentien, D.; Pierron, G.; Lapouble, E.; Bellini, A.; Clement, N.; Iacono, I.; et al. Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma. Clin. Cancer Res. 2016, 22, 5564–5573. [Google Scholar] [CrossRef]
- Singh, A.D.; Topham, A. Incidence of Uveal Melanoma in the United States: 1973–1997. Ophthalmology 2003, 110, 956–961. [Google Scholar] [CrossRef]
- Damato, B.; Groenewald, C.; Mcgalliard, J.; Wong, D. Endoresection of choroidal melanoma. Br. J. Ophthalmol. 1998, 82, 213–218. [Google Scholar] [CrossRef]




| Gender | Values |
|---|---|
| Female, No. (%) | 11 (46%) |
| Male, No. (%) | 13 (54%) |
| Age, years | 50.2 ± 13.8 (20.8, 73.9) |
| Tumor Characteristics at Diagnosis | |
| Thickness, mm | 8.6 ± 1.7 (4.8, 12.3) |
| Largest basal diameter, mm | 12.4 ± 2.3 (8.3, 15.7) |
| Tumor Stage (8th TNM Classification [23]) | |
| T1, No. (%) | 0 (0%) |
| T2, No. (%) | 7 (29%) |
| T3, No. (%) | 17 (71%) |
| T4, No. (%) | 0 (0%) |
| Duration between Treatment Events, Month | |
| Fine-needle biopsy to proton beam irradiation | 0.5 ± 0.1 (0.4, 0.7) |
| Proton therapy to endoresection | 1.9 ± 0.5 (1.3, 3.0) |
| Fine-needle biopsy to endoresection | 2.4 ± 0.5 (1.9, 3.5) |
| Follow-up | |
| Follow-up after diagnosis, years | 3.7 ± 1.6 (0.7, 5.8) |
| Secondary enucleation, No. (%) | 3 (13%) |
| Metastasis development, No. (%) | 3 (13%) |
| Death, No. (%) | 1 (4%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matet, A.; Aït Raïs, K.; Malaise, D.; Angi, M.; Dendale, R.; Tick, S.; Lumbroso-Le Rouic, L.; Lévy-Gabriel, C.; Rodrigues, M.; Pierron, G.; et al. Comparative Cytogenetic Abnormalities in Paired Choroidal Melanoma Samples Obtained Before and After Proton Beam Irradiation by Transscleral Fine-Needle Aspiration Biopsy and Endoresection. Cancers 2019, 11, 1173. https://doi.org/10.3390/cancers11081173
Matet A, Aït Raïs K, Malaise D, Angi M, Dendale R, Tick S, Lumbroso-Le Rouic L, Lévy-Gabriel C, Rodrigues M, Pierron G, et al. Comparative Cytogenetic Abnormalities in Paired Choroidal Melanoma Samples Obtained Before and After Proton Beam Irradiation by Transscleral Fine-Needle Aspiration Biopsy and Endoresection. Cancers. 2019; 11(8):1173. https://doi.org/10.3390/cancers11081173
Chicago/Turabian StyleMatet, Alexandre, Khadija Aït Raïs, Denis Malaise, Martina Angi, Rémi Dendale, Sarah Tick, Livia Lumbroso-Le Rouic, Christine Lévy-Gabriel, Manuel Rodrigues, Gaëlle Pierron, and et al. 2019. "Comparative Cytogenetic Abnormalities in Paired Choroidal Melanoma Samples Obtained Before and After Proton Beam Irradiation by Transscleral Fine-Needle Aspiration Biopsy and Endoresection" Cancers 11, no. 8: 1173. https://doi.org/10.3390/cancers11081173
APA StyleMatet, A., Aït Raïs, K., Malaise, D., Angi, M., Dendale, R., Tick, S., Lumbroso-Le Rouic, L., Lévy-Gabriel, C., Rodrigues, M., Pierron, G., & Cassoux, N. (2019). Comparative Cytogenetic Abnormalities in Paired Choroidal Melanoma Samples Obtained Before and After Proton Beam Irradiation by Transscleral Fine-Needle Aspiration Biopsy and Endoresection. Cancers, 11(8), 1173. https://doi.org/10.3390/cancers11081173