Tumor Transcriptome Reveals High Expression of IL-8 in Non-Small Cell Lung Cancer Patients with Low Pectoralis Muscle Area and Reduced Survival
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. PMA Distinguishes NSCLC Patients with Low- and High-Muscularity
2.3. Patients with Low-Muscularity Upregulate Tumor Genes Previously Associated with Cachexia
2.4. Secretome-Related Genes with Prognostic Value in NSCLC
2.5. High IL-8 Expression in Tumor Tissues is Associated with Poor Prognosis in NSCLC
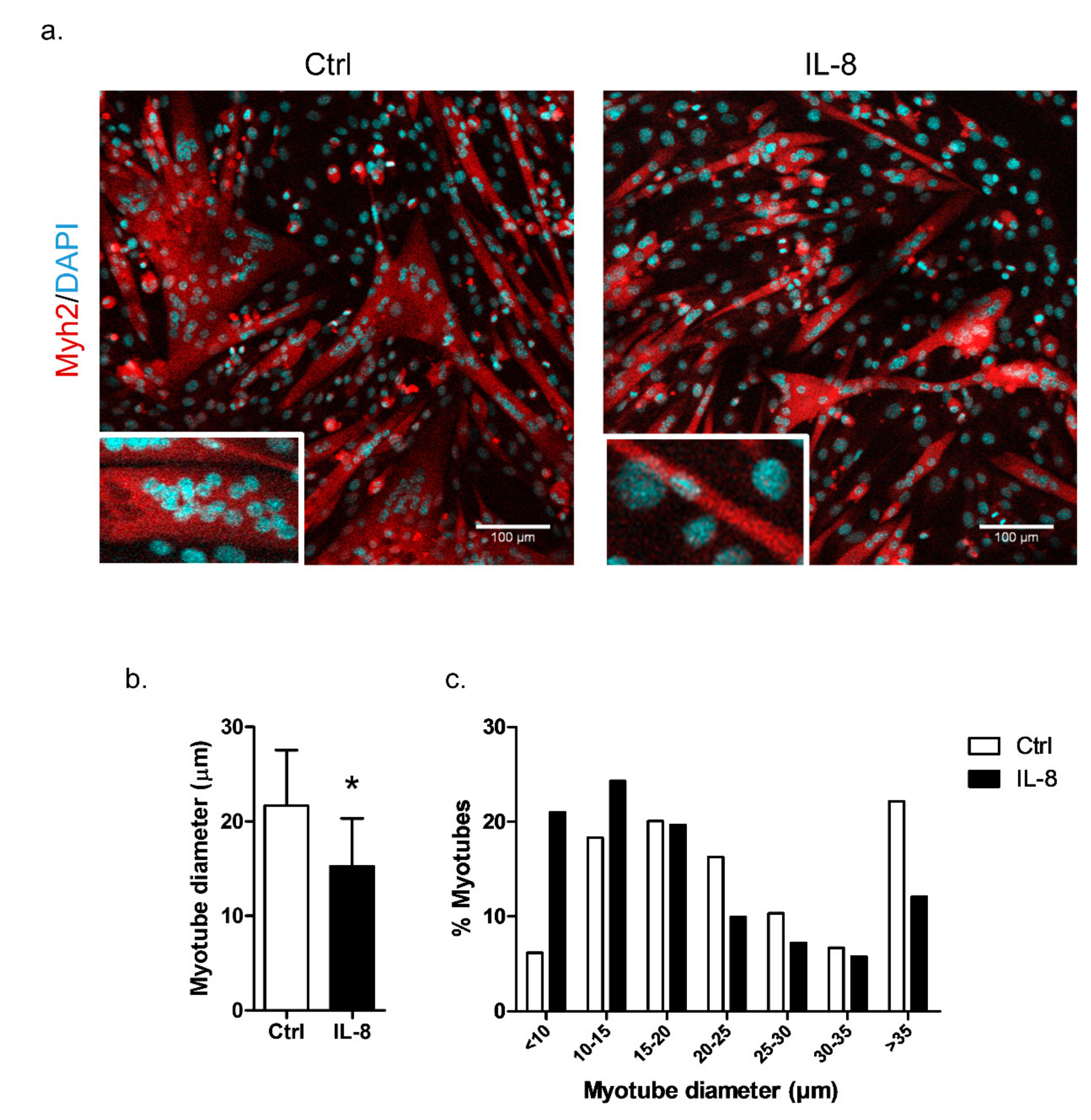
2.6. IL-8 Treatment Induces In Vitro Myotube Atrophy
3. Discussion
4. Materials and Methods
4.1. Datasets
4.2. CT Imaging Analyses
4.3. Gene Expression Analysis
4.4. Gene Ontology Enrichment Analysis
4.5. Protein-Protein Interactions (PPI) Networks
4.6. In Silico Identification of Secreted Proteins
4.7. Prognostic Performance of Secretory Genes in Predicting NSCLC Outcome
4.8. Functional Assay Using the C2C12 Cell Culture
4.9. Immunofluorescence Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nat. Publ. Gr. 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Mitchell, C. Lung cancer. Aust. Fam. Physician 2004, 33, 321–325. [Google Scholar] [PubMed]
- Baracos, V.E.; Reiman, T.; Mourtzakis, M.; Gioulbasanis, I.; Antoun, S. Body composition in patients with non-small cell lung cancer: A contemporary view of cancer cachexia with the use of computed tomography image analysis. Am. J. Clin. Nutr. 2010, 91, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Mytelka, D.S.; Li, L.; Benoit, K. Post-diagnosis weight loss as a prognostic factor in non-small cell lung cancer. J. Cachexia. Sarcopenia Muscle 2018, 9, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Unit, L.; Hospital, R.M.; Road, D. Do patients with weight loss have a worse outcome when undergoing chemotherapy for lung cancers? Br. J. Cancer 2004, 90, 1905–1911. [Google Scholar]
- Dahlberg, S.E.; Schiller, J.H.; Bonomi, P.B.; Sandler, A.B.; Brahmer, J.R.; Ramalingam, S.S.; Johnson, D.H. Body mass index and its association with clinical outcomes for advanced non-small-cell lung cancer patients enrolled on eastern cooperative oncology group clinical trials. J. Thorac. Oncol. 2013, 8, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Birdsell, L.; MacDonald, N.; Reiman, T.; Clandinin, M.T.; McCargar, L.J.; Murphy, R.; Ghosh, S.; Sawyer, M.B.; Baracos, V.E. Cancer cachexia in the age of obesity: Skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J. Clin. Oncol. 2013, 31, 1539–1547. [Google Scholar] [CrossRef]
- Stene, G.B.; Helbostad, J.L.; Amundsen, T.; Sørhaug, S.; Hjelde, H.; Kaasa, S.; Grønberg, B.H. Changes in skeletal muscle mass during palliative chemotherapy in patients with advanced lung cancer. Acta Oncol. 2015, 54, 340–348. [Google Scholar] [CrossRef]
- Prado, C.M.M.; Lieff, J.R.; Mccargar, L.J.; Reiman, T.; Sawyer, M.B.; Martin, L.; Baracos, V.E. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: A population-based study. Lancet Oncol. 2008, 9, 629–635. [Google Scholar] [CrossRef]
- Management, S.; Care, S.; Assessment, G. Nutritional Status, Body Surface, and Low Lean Body Mass/Body Mass Index Are Related to Dose Reduction and Severe Gastrointestinal Toxicity Induced by Afatinib in Patients with Non-Small Cell Lung Cancer. Oncologist 2015, 20, 967–974. [Google Scholar]
- Sjøblom, B.; Grønberg, B.H.; Benth, J.Š.; Baracos, V.E.; Øystein, F.; Hjermstad, M.J.; Aass, N.; Jordhøy, M. Low muscle mass is associated with chemotherapy-induced haematological toxicity in advanced non-small cell lung cancer. Lung Cancer 2015, 90, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, C.M.; San Jose Estepar, R.; van der Velden, J.; Cole, B.F.; Christiani, D.C.; Washko, G.R.; Cancer, C.L.; van der Velden, J.; Cole, B.F.; Kinsey, C.M.; et al. Lower Pectoralis Muscle Area Is Associated with a Worse Overall Survival in Non-Small Cell Lung Cancer. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.; Sjøblom, B.; Wentzel-Larsen, T.; Grønberg, B.H.; Baracos, V.E.; Hjermstad, M.J.; Aass, N.; Bremnes, R.M.; Fløtten, Ø.; Jordhøy, M. Muscle mass and association to quality of life in non-small cell lung cancer patients. J. Cachexia. Sarcopenia Muscle 2017, 8, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Kilgour, R.D.; Vigano, A.; Trutschnigg, B.; Hornby, L.; Lucar, E.; Bacon, S.L.; Morais, J.A. Cancer-related fatigue: The impact of skeletal muscle mass and strength in patients with advanced cancer. J. Cachexia. Sarcopenia Muscle 2010, 1, 177–185. [Google Scholar] [CrossRef]
- Go, S.; Park, M.J.; Song, H.; Kang, M.H.; Park, H.J.; Jeon, K.N.; Kim, S.; Kim, M.J.; Kang, J.; Lee, G. Sarcopenia and inflammation are independent predictors of survival in male patients newly diagnosed with small cell lung cancer. Support. Care Cancer 2016, 24, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Tsoli, M.; Robertson, G. Cancer cachexia: malignant inflammation, tumorkines, and metabolic mayhem. Trends Endocrinol. Metab. 2013, 24, 174–183. [Google Scholar] [CrossRef]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Publ. Gr. 2018, 4, 1–18. [Google Scholar]
- Twelkmeyer, B.; Tardif, N.; Rooyackers, O. Omics and cachexia. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 181–185. [Google Scholar] [CrossRef]
- Hsiao, Y.; Chu, L.; Chen, J.; Yeh, T.; Yu, J. Proteomic profiling of the cancer cell secretome: Informing clinical research. Expert Rev. Proteomics 2017, 14, 737–756. [Google Scholar] [CrossRef] [PubMed]
- Schaaij-visser, T.B.M.; de Wit, M.; Lam, S.W.; Jiménez, C.R. The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context. BBA Proteins Proteomics 2013, 1834, 2242–2258. [Google Scholar] [CrossRef] [PubMed]
- Pavlou, M.P.; Diamandis, E.P. The cancer cell secretome: A good source for discovering biomarkers? J. Proteom. 2010, 73, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Fukawa, T.; Yan-Jiang, B.C.; Min-Wen, J.C.; Jun-Hao, E.T.; Huang, D.; Qian, C.-N.; Ong, P.; Li, Z.; Chen, S.; Mak, S.Y.; et al. Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia. Nat. Med. 2016, 22, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Oeing, C.U.; Rohm, M.; Baysal-Temel, E.; Lehmann, L.H.; Bauer, R.; Volz, H.C.; Boutros, M.; Sohn, D.; Sticht, C.; et al. Ataxin-10 is part of a cachexokine cocktail triggering cardiac metabolic dysfunction in cancer cachexia. Mol. Metab. 2016, 5, 67–78. [Google Scholar] [CrossRef] [PubMed]
- McLean, J.B.; Moylan, J.S.; Horrell, E.M.W.; Andrade, F.H. Proteomic analysis of media from lung cancer cells reveals role of 14-3-3 proteins in cachexia. Front. Physiol. 2015, 6, 1–8. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Rios Velazquez, E.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Lambin, P. Data From NSCLC-Radiomics-Genomics. Cancer Imaging Arch. 2015. [Google Scholar] [CrossRef]
- Penafuerte, C.A.; Gagnon, B.; Sirois, J.; Murphy, J.; Macdonald, N.; Tremblay, M.L. Identification of neutrophil-derived proteases and angiotensin II as biomarkers of cancer cachexia. Br. J. Cancer 2016, 114, 680–687. [Google Scholar] [CrossRef]
- Kuroda, K.; Nakashima, J.; Kanao, K.; Kikuchi, E.; Miyajima, A.; Horiguchi, Y.; Nakagawa, K.; Oya, M.; Ohigashi, T.; Murai, M. Interleukin 6 is associated with cachexia in patients with prostate cancer. Urology 2007, 69, 113–117. [Google Scholar] [CrossRef]
- Hou, Y.-C.; Wang, C.-J.; Chao, Y.-J.; Chen, H.-Y.; Wang, H.-C.; Tung, H.-L.; Lin, J.-T.; Shan, Y.-S. Elevated Serum Interleukin-8 Level Correlates with Cancer-Related Cachexia and Sarcopenia: An Indicator for Pancreatic Cancer Outcomes. J. Clin. Med. 2018, 7, 502. [Google Scholar] [CrossRef] [PubMed]
- Kandarian, S.C.; Nosacka, R.L.; Delitto, A.E.; Judge, A.R.; Judge, S.M.; Ganey, J.D.; Moreira, J.D.; Jackman, R.W. Tumour-derived leukaemia inhibitory factor is a major driver of cancer cachexia and morbidity in C26 tumour-bearing mice. J. Cachexia. Sarcopenia Muscle 2018, 9, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Carson, J.A.; Baltgalvis, K.A. Interleukin 6 as a key regulator of muscle mass during cachexia. Exerc. Sport Sci. Rev. 2010, 38, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.M.; Mariano, V.S.; Aguiar Pastrez, P.R.; Pinto, M.C.; Castro, A.G.; Syrjanen, K.J.; Longatto-Filho, A. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PLoS ONE 2017, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, K.; Andersen, S.; Degen, S.; Tadini, V.; Grosjean, J.; Hatakeyama, S.; Tesfahun, A.N.; Moestue, S.; Kim, J.; Nonstad, U.; et al. Cancer cachexia associates with a systemic autophagy-inducing activity mimicked by cancer cell-derived IL-6 trans-signaling. Sci. Rep. 2017, 7, 2046. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.; Diaz, A.; Ross, J.; San Jose Estepar, R.; Zhou, L.; Washko, G. Quantitative computed tomography measures of pectoralis muscle area and disease severity in chronic obstructive pulmonary disease. A cross-sectional study. Ann. Am. Thorac. Soc. 2014, 11, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Teigen, L.; John, R.; Kuchnia, A.; Nage, E.; Earthman, C.; Kealhofer, J.; Martin, C.; Cogswell, R. Preoperative Pectoralis Muscle Quantity and Attenuation by Computed Tomography Are Novel and Powerful Predictors of Mortality After Left Ventricular Assist Device Implantation. Circ. Hear. Fail. 2017, 10, e004069. [Google Scholar] [CrossRef]
- Fearon, K.C.H.; Glass, D.J.; Guttridge, D.C. Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell Metab. 2012, 16, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Jafri, R.; Previgliano, C.; Khandelwal, K.; Shi, R. Cachexia Index in Advanced Non-Small-Cell Lung Cancer Patients. Clin. Med. Insights Oncol. 2015, 9, 87–93. [Google Scholar] [CrossRef]
- Srdic, D.; Plestina, S.; Sverko-Peternac, A.; Nikolac, N.; Simundic, A.M.; Samarzija, M. Cancer cachexia, sarcopenia and biochemical markers in patients with advanced non-small cell lung cancer—chemotherapy toxicity and prognostic value. Support. Care Cancer 2016, 24, 4495–4502. [Google Scholar] [CrossRef] [PubMed]
- Pfitzenmaier, J.; Vessella, R.; Higano, C.S.; Noteboom, J.L.; Wallace, D.; Corey, E. Elevation of cytokine levels in cachectic patients with prostate carcinoma. Cancer 2003, 97, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Tazaki, E.; Shimizu, N.; Tanaka, R.; Yoshizumi, M.; Kamma, H.; Imoto, S.; Goya, T.; Kozawa, K.; Nishina, A.; Kimura, H. Serum cytokine profiles in patients with prostate carcinoma. Exp. Ther. Med. 2011, 2, 887–891. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.J.; Figuerêdo, R.G.; Azevedo, F.F.; Cavallaro, D.A.; Neto, N.I.P.; Lima, J.D.C.; Matos-Neto, E.; Radloff, K.; Riccardi, D.M.; Camargo, R.G.; et al. Adipose tissue fibrosis in human cancer cachexia: The role of TGFβ pathway. BMC Cancer 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Zhang, D.; Song, B.; Wang, S.; Zheng, H.; Wang, X. Association of interleukin-8 with cachexia from patients with low-third gastric cancer. Comp. Funct. Genomics 2009, 2009, 1–6. [Google Scholar]
- Lerner, L.; Hayes, T.G.; Tao, N.; Krieger, B.; Feng, B.; Wu, Z.; Nicoletti, R.; Isabel Chiu, M.; Gyuris, J.; Garcia, J.M. Plasma growth differentiation factor 15 is associated with weight loss and mortality in cancer patients. J. Cachexia. Sarcopenia Muscle 2015, 6, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Richey, L.M.; George, J.R.; Couch, M.E.; Kanapkey, B.K.; Yin, X.; Cannon, T.; Stewart, P.W.; Weissler, M.C.; Shores, C.G. Defining cancer cachexia in head and neck squamous cell carcinoma. Clin. Cancer Res. 2007, 13, 6561–6567. [Google Scholar] [CrossRef] [PubMed]
- Krzystek-Korpacka, M.; Matusiewicz, M.; Diakowska, D.; Grabowski, K.; Blachut, K.; Kustrzeba-Wojcicka, I.; Banas, T. Impact of weight loss on circulating IL-1, IL-6, IL-8, TNF-α, VEGF-A, VEGF–C and midkine in gastroesophageal cancer patients. Clin. Biochem. 2007, 40, 1353–1360. [Google Scholar] [CrossRef]
- Dolan, R.D.; Almasaudi, A.S.; Dieu, L.B.; Horgan, P.G.; McSorley, S.T.; McMillan, D.C. The relationship between computed tomography-derived body composition, systemic inflammatory response, and survival in patients undergoing surgery for colorectal cancer. J. Cachex Sarcopenia Muscle 2018, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, D.P.J.; Krill, M.; Farshidfar, F.; Li, T.; Rensen, S.S.; Olde Damink, S.W.M.; Dixon, E.; Sutherland, F.R.; Ball, C.G.; Mazurak, V.C.; et al. Host phenotype is associated with reduced survival independent of tumour biology in patients with colorectal liver metastases. J. Cachexia. Sarcopenia Muscle 2018, 10, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, C.A.N.; Salafranca, M.N.; Adhikari, S.; Xia, Y.; Feng, L.; Harrison, J.K. Identification of two rat genes orthologous to the human interleukin-8 receptors. J. Biol. Chem. 1996, 271, 32770–32776. [Google Scholar] [CrossRef]
- Gerber, M.H.; Underwood, P.W.; Judge, S.M.; Delitto, D.; Delitto, A.E.; Nosacka, R.L.; DiVita, B.B.; Thomas, R.M.; Permuth, J.B.; Hughes, S.J.; et al. Local and Systemic Cytokine Profiling for Pancreatic Ductal Adenocarcinoma to Study Cancer Cachexia in an Era of Precision Medicine. Int. J. Mol. Sci. 2018, 19, 3836. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Freymann, J.; Kirby, J.; Perry, J.; Clunie, D.; Jaffe, C. Image data sharing for biomedical research--meeting HIPAA requirements for De-identification. J. Digit. Imaging 2012, 25, 14–24. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kim, Y.S.; Park, I.; Ahn, H.K. Evaluation of sarcopenia in small-cell lung cancer patients by routine chest CT. Support. Care Cancer 2016, 24, 4721–4726. [Google Scholar] [CrossRef]
- Nilufer, G.; Ayse, Y. Estimation of stature and sex from sternal lengths: an autopsy study. Anat. Sci. Int. 2015, 90, 89–96. [Google Scholar]
- Zhou, S.H.; McCarthy, I.D.; McGregor, A.H.; Coombs, R.R.H.; Hughes, S.P.F. Geometrical dimensions of the lower lumbar vertebrae-analysis of data from digitised CT images. Eur. Spine J. 2000, 9, 242–248. [Google Scholar] [CrossRef]
- Yaguchi, Y.; Kumata, Y.; Horikawa, M.; Kiyokawa, T.; Iinuma, H.; Inaba, T.; Fukushima, R. Clinical Significance of Area of Psoas Major Muscle on Computed Tomography after Gastrectomy in Gastric Cancer Patients. Ann. Nutr. Metab. 2017, 71, 145–149. [Google Scholar] [CrossRef]
- Blake, J.; Christie, K.; Dolan, M.; Drabkin, H.; Hill, D.; Ni, L.; Sitnikov, D.; Westerfield, M. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015, 43, 1049–1056. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Petersen, T.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011, 29, 785–786. [Google Scholar] [CrossRef]
- Bendtsen, J.; Jensen, L.; Blom, N.; von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.; Reid, G.; Simpson, R. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, 1241–1244. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Feizi, A.; Banaei-esfahani, A.; Nielsen, J. Database tool HCSD: The human cancer secretome database. Database (Oxford) 2015, 2015, bav051. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.; Kim, S.; Choi, D.; Yoon, Y. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar]
- Nanjappa, V.; Thomas, J.; Marimuthu, A.; Muthusamy, B. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, 959–965. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Rousseaux, S.; Debernardi, A.; Jacquiau, B.; Vitte, A. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 2013, 5, 186ra66. [Google Scholar] [CrossRef]
- Yamauchi, M.; Yamaguchi, R.; Nakata, A.; Kohno, T. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS ONE 2012, 7, e43923. [Google Scholar] [CrossRef]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef]
- Shedden, K.; Taylor, J.; Enkemann, S. Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 2008, 14, 822–827. [Google Scholar]
- Lee, E.; Son, D.; Kim, S.; Lee, J. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin. Cancer Res. 2008, 14, 7397–7404. [Google Scholar] [CrossRef]
- Surowiak, P.; Budczies, J. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PLoS ONE 2013, 8, e82241. [Google Scholar]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef]





| Characteristics | All | Men | Women | p-value * |
|---|---|---|---|---|
| Number of patients | 89 | 60 | 29 | |
| Age | 65.2 ± 8.7 | 66.9 ± 7.3 | 61.8 ± 10.3 | 0.011 a |
| Cancer Stage (%) | ||||
| Early Stages (I-II) | 79.7 | 74.6 | 90 | 0.08 b |
| Advanced Stages (III-IV) | 20.2 | 25.4 | 10 | |
| Histological Type (%) | ||||
| Adenocarcinoma | 47.2 | 37.3 | 66.6 | 0.02 b |
| Squamous Cell Carcinoma | 40.4 | 49.2 | 23.3 | |
| Other Subtypes | 12.4 | 13.5 | 10.1 | |
| PMA (cm2) | 37.3 ± 11.1 | 42.5 ± 9.3 | 27 ± 6.0 | <0.001 a |
| HM (N) | 59 | 40 | 19 | |
| LM (N) | 30 | 20 | 10 | |
| LM PMA (cm2) | 28.6 ± 6.5 #,c | 32.3 ± 4.3 #,c | 21 ± 1.4 #,c | <0.001 a |
| HM PMA (cm2) | 41.7 ± 10.2 | 47.5 ± 6.3 | 30 ± 5.2 | <0.001 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiloni Cury, S.; de Moraes, D.; Paccielli Freire, P.; de Oliveira, G.; Venâncio Pereira Marques, D.; Javier Fernandez, G.; Dal-Pai-Silva, M.; Nishida Hasimoto, É.; Pintor dos Reis, P.; Regina Rogatto, S.; et al. Tumor Transcriptome Reveals High Expression of IL-8 in Non-Small Cell Lung Cancer Patients with Low Pectoralis Muscle Area and Reduced Survival. Cancers 2019, 11, 1251. https://doi.org/10.3390/cancers11091251
Santiloni Cury S, de Moraes D, Paccielli Freire P, de Oliveira G, Venâncio Pereira Marques D, Javier Fernandez G, Dal-Pai-Silva M, Nishida Hasimoto É, Pintor dos Reis P, Regina Rogatto S, et al. Tumor Transcriptome Reveals High Expression of IL-8 in Non-Small Cell Lung Cancer Patients with Low Pectoralis Muscle Area and Reduced Survival. Cancers. 2019; 11(9):1251. https://doi.org/10.3390/cancers11091251
Chicago/Turabian StyleSantiloni Cury, Sarah, Diogo de Moraes, Paula Paccielli Freire, Grasieli de Oliveira, Douglas Venâncio Pereira Marques, Geysson Javier Fernandez, Maeli Dal-Pai-Silva, Érica Nishida Hasimoto, Patricia Pintor dos Reis, Silvia Regina Rogatto, and et al. 2019. "Tumor Transcriptome Reveals High Expression of IL-8 in Non-Small Cell Lung Cancer Patients with Low Pectoralis Muscle Area and Reduced Survival" Cancers 11, no. 9: 1251. https://doi.org/10.3390/cancers11091251
APA StyleSantiloni Cury, S., de Moraes, D., Paccielli Freire, P., de Oliveira, G., Venâncio Pereira Marques, D., Javier Fernandez, G., Dal-Pai-Silva, M., Nishida Hasimoto, É., Pintor dos Reis, P., Regina Rogatto, S., & Francisco Carvalho, R. (2019). Tumor Transcriptome Reveals High Expression of IL-8 in Non-Small Cell Lung Cancer Patients with Low Pectoralis Muscle Area and Reduced Survival. Cancers, 11(9), 1251. https://doi.org/10.3390/cancers11091251







