MicroRNAs and Metastasis
Abstract
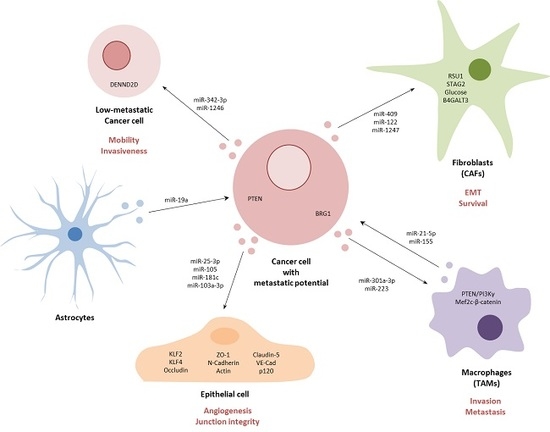
:
Graphical Abstract
1. Introduction
2. Metastasis—Promoting miRNAs
3. Metastasis—Suppressing miRNAs
4. Metastasis and Circulating miRNAs
5. The Metastatic Targetome
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Winter, M.; Gibson, R.; Ruszkiewicz, A.; Thompson, S.K.; Thierry, B. Beyond conventional pathology: Towards preoperative and intraoperative lymph node staging. Int. J. Cancer 2015, 136, 743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.; Fodde, R. Cancer stemness and metastasis: Therapeutic consequences and perspectives. Eur. J. Cancer 2010, 46, 1198. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92. [Google Scholar] [CrossRef] [Green Version]
- Lawrie, C.H. MicroRNAs in Medicine; Wiley: New York, NY, USA, 2014. [Google Scholar]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Gangoda, L.; Boukouris, S.; Liem, M.; Kalra, H.; Mathivanan, S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics 2015, 15, 260. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.J.; Azhar, S.; Kraemer, F.B. SR-B1: A Unique Multifunctional Receptor for Cholesterol Influx and Efflux. Annu. Rev. Physiol. 2018, 80, 95. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs—The micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293. [Google Scholar] [CrossRef]
- Ma, L.; Weinberg, R.A. Micromanagers of malignancy: Role of microRNAs in regulating metastasis. Trends Genet. Genet. 2008, 24, 448. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.O.; Kwak, S.Y.; An, H.J.; Bae, I.H.; Park, M.J.; Han, Y.H. miR-181b-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization by directly targeting YWHAG. Biochim. Biophys. Acta 2016, 1863 Pt 7 A, 1601. [Google Scholar] [CrossRef]
- Cai, J.; Guan, H.; Fang, L.; Yang, Y.; Zhu, X.; Yuan, J.; Wu, J.; Li, M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J. Clin. Investig. 2013, 123, 566. [Google Scholar]
- Chen, Y.; Zhang, J.; Wang, H.; Zhao, J.; Xu, C.; Du, Y.; Luo, X.; Zheng, F.; Liu, R.; Zhang, H.; et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer 2012, 12, 111. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.D.; Xin, H.N.; Yang, Z.C.; Wang, W.J.; Dong, J.J.; Jin, L.Y.; Li, F.N. miR-135b promotes proliferation and metastasis by targeting APC in triple-negative breast cancer. J. Cell. Physiol. 2019, 234, 10819. [Google Scholar] [CrossRef]
- Zeng, Y.B.; Liang, X.H.; Zhang, G.X.; Jiang, N.; Zhang, T.; Huang, J.Y.; Zhang, L.; Zeng, X.C. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016, 16, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Y.; Liang, H.; Uzair Ur, R.; Wang, Y.; Zhang, W.; Zhou, Y.; Chen, S.; Yu, M.; Cui, S.; Liu, M.; et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 2016, 6, 37421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, T.; Yang, Z.; Zhu, Q.; Wu, Y.; Sun, K.; Alahdal, M.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 2018, 32, 6965–6981. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a contributes to breast cancer cells epithelial-mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, M.; Huang, J.; Li, Y.; Wang, S.; Harrington, C.A.; Qian, D.Z.; Sun, X.X.; Dai, M.S. microRNA-130a suppresses breast cancer cell migration and invasion by targeting FOSL1 and upregulating ZO-1. J. Cell. Biochem. 2018, 119, 4945. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, H.; Gao, C.; Lv, Y.; Chen, X.; Xu, R.; Sun, M.; Liu, X.; Lu, X.; Pei, X.; et al. Tumor-promoting properties of miR-8084 in breast cancer through enhancing proliferation, suppressing apoptosis and inducing epithelial-mesenchymal transition. J. Transl. Med. 2018, 16, 38. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Bi, L.; Wang, Q.; Wen, M.; Li, C.; Ren, Y.; Jiao, Q.; Mao, J.H.; Wang, C.; Wei, G.; et al. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene 2018, 37, 3426. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chen, X.; Sun, K.X.; Xiu, Y.L.; Liu, B.L.; Feng, M.X.; Sang, X.B.; Zhao, Y. MicroRNA-93 Promotes Epithelial-Mesenchymal Transition of Endometrial Carcinoma Cells. PLoS ONE 2016, 11, e0165776. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Wang, P.; Lin, J.; Zheng, X.; Yang, F.; Zhang, G.; Chen, D.; Xie, J.; Gao, Z.; Peng, L.; et al. MicroRNA-197 Promotes Metastasis of Hepatocellular Carcinoma by Activating Wnt/beta-Catenin Signaling. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharm. 2018, 51, 470. [Google Scholar] [CrossRef]
- Luo, Y.; Wu, J.; Wu, Q.; Li, X.; Wu, J.; Zhang, J.; Rong, X.; Rao, J.; Liao, Y.; Bin, J.; et al. miR-577 Regulates TGF-beta Induced Cancer Progression through a SDPR-Modulated Positive-Feedback Loop with ERK-NF-kappaB in Gastric Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2019. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.R.; Wen, L.Q.; Wang, Y.; Zhou, T.C.; Ma, N.; Hou, Z.H.; Jiang, Z.P. MicroRNA-520c enhances cell proliferation, migration, and invasion by suppressing IRF2 in gastric cancer. FEBS Openbio 2016, 6, 1257. [Google Scholar] [CrossRef]
- Bu, P.; Wang, L.; Chen, K.Y.; Rakhilin, N.; Sun, J.; Closa, A.; Tung, K.L.; King, S.; Kristine Varanko, A.; Xu, Y.; et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta. Nat. Commun. 2015, 6, 6879. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Yang, J.; Xiao, B. MicroRNA-20b (miR-20b) Promotes the Proliferation, Migration, Invasion, and Tumorigenicity in Esophageal Cancer Cells via the Regulation of Phosphatase and Tensin Homologue Expression. PLoS ONE 2016, 11, e0164105. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Zhu, Y.; Dai, Y.; Zeng, T.; Liu, L.; Li, J.; Wang, H.; Qin, Y.; Zeng, M.; et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 2014, 5, 11669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Zhou, X.; Yin, Z.; Guo, J.; Hu, T.; Jiang, S.; Liu, L.; Dong, X.; Zhang, S.; Wu, G. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci. Rep. 2016, 6, 35714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, T.; Li, L.; Cheng, Y.; Ren, C.; Zhang, G. MicroRNA-194 promotes the growth, migration, and invasion of ovarian carcinoma cells by targeting protein tyrosine phosphatase nonreceptor type 12. Onco Targets Ther. 2016, 9, 4307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846. [Google Scholar] [CrossRef] [Green Version]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593. [Google Scholar] [CrossRef]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910. [Google Scholar] [CrossRef] [Green Version]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Liu, J.; Li, X.; Xiao, G.; Wang, H.; Yang, G.; Li, Y.; Tang, S.C.; Qin, S.; Du, N.; et al. MYC and DNMT3A-mediated DNA methylation represses microRNA-200b in triple negative breast cancer. J. Cell. Mol. Med. 2018, 22, 6262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y.; et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Yamamoto, N.; Yoshino, H.; Itesako, T.; Enokida, H.; Nakagawa, M.; Okamoto, Y.; et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br. J. Cancer 2013, 109, 2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukumoto, I.; Kikkawa, N.; Matsushita, R.; Kato, M.; Kurozumi, A.; Nishikawa, R.; Goto, Y.; Koshizuka, K.; Hanazawa, T.; Enokida, H.; et al. Tumor-suppressive microRNAs (miR-26a/b, miR-29a/b/c and miR-218) concertedly suppressed metastasis-promoting LOXL2 in head and neck squamous cell carcinoma. J. Hum. Genet. 2016, 61, 109. [Google Scholar] [CrossRef]
- Nishikawa, R.; Chiyomaru, T.; Enokida, H.; Inoguchi, S.; Ishihara, T.; Matsushita, R.; Goto, Y.; Fukumoto, I.; Nakagawa, M.; Seki, N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. Febs Lett. 2015, 589, 2136. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.F.; Zhang, J.G.; Guo, T.X.; Pan, X.J. MiR-504 inhibits cell proliferation and invasion by targeting LOXL2 in non small cell lung cancer. Biomed. Pharm. 2018, 97, 1289. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Zhao, M.; Wu, C.; Zhang, P.; Tang, L.; Zhang, H.; Chen, X.; Yang, Y.; Liu, G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin beta1 and matrix metalloproteinase2 (MMP2). PLoS ONE 2013, 8, e70192. [Google Scholar]
- Benaich, N.; Woodhouse, S.; Goldie, S.J.; Mishra, A.; Quist, S.R.; Watt, F.M. Rewiring of an epithelial differentiation factor, miR-203, to inhibit human squamous cell carcinoma metastasis. Cell Rep. 2014, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Park, S.I.; McCauley, L.K.; Wang, C.Y. Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013, 288, 10241. [Google Scholar] [CrossRef] [Green Version]
- Lohcharoenkal, W.; Das Mahapatra, K.; Pasquali, L.; Crudden, C.; Kular, L.; Akkaya Ulum, Y.Z.; Zhang, L.; Xu Landen, N.; Girnita, L.; Jagodic, M.; et al. Genome-Wide Screen for MicroRNAs Reveals a Role for miR-203 in Melanoma Metastasis. J. Investig. Dermatol. 2018, 138, 882. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, X.; Zhao, G.; Yan, H.; Dong, P.; Watari, H.; Sims, M.; Li, W.; Pfeffer, L.M.; Guo, Y.; et al. miR-203 inhibits ovarian tumor metastasis by targeting BIRC5 and attenuating the TGFbeta pathway. J. Exp. Clin. Cancer Res. 2018, 37, 235. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, B.; Cui, J.; Liang, Z.; Liu, K. miR-203 inhibits the invasion and EMT of gastric cancer cells by directly targeting Annexin A4. Oncol. Res. 2019. [Google Scholar] [CrossRef]
- Deng, B.; Wang, B.; Fang, J.; Zhu, X.; Cao, Z.; Lin, Q.; Zhou, L.; Sun, X. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci. Rep. 2016, 6, 28301. [Google Scholar] [CrossRef] [Green Version]
- Xi, Z.W.; Xin, S.Y.; Zhou, L.Q.; Yuan, H.X.; Wang, Q.; Chen, K.X. Downregulation of rho-associated protein kinase 1 by miR-124 in colorectal cancer. World J. Gastroenterol. 2015, 21, 5454. [Google Scholar] [CrossRef]
- Shin, J.Y.; Kim, Y.I.; Cho, S.J.; Lee, M.K.; Kook, M.C.; Lee, J.H.; Lee, S.S.; Ashktorab, H.; Smoot, D.T.; Ryu, K.W.; et al. MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. PLoS ONE 2014, 9, e85205. [Google Scholar] [CrossRef]
- Mohammadi-Yeganeh, S.; Hosseini, V.; Paryan, M. Wnt pathway targeting reduces triple-negative breast cancer aggressiveness through miRNA regulation in vitro and in vivo. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Kong, X.; Li, G.; Yuan, Y.; He, Y.; Wu, X.; Zhang, W.; Wu, Z.; Chen, T.; Wu, W.; Lobie, P.E.; et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE 2012, 7, e41523. [Google Scholar] [CrossRef]
- Sossey-Alaoui, K.; Downs-Kelly, E.; Das, M.; Izem, L.; Tubbs, R.; Plow, E.F. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int. J. Cancer 2011, 129, 1331. [Google Scholar] [CrossRef] [Green Version]
- Augoff, K.; Das, M.; Bialkowska, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. miR-31 is a broad regulator of beta1-integrin expression and function in cancer cells. Mol. Cancer Res. 2011, 9, 1500. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Fei, D.; Zong, S.; Fan, Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am. J. Transl. Res. 2016, 8, 2620. [Google Scholar]
- Tang, W.; Xu, P.; Wang, H.; Niu, Z.; Zhu, D.; Lin, Q.; Tang, L.; Ren, L. MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2. Onco Targets Ther. 2018, 11, 2319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, W.L.; Huang, W.D.; Li, B.; Chen, T.R.; Li, Z.X.; Zhao, C.L.; Li, H.Y.; Wu, Y.M.; Yan, W.J.; Xiao, J.R. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol. Cancer 2018, 17, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramchandani, D.; Lee, S.K.; Yomtoubian, S.; Han, M.S.; Tung, C.H.; Mittal, V. Nanoparticle Delivery of miR-708 Mimetic Impairs Breast Cancer Metastasis. Mol. Cancer Ther. 2019, 18, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Liu, A.Y.; Fan, C.; Zheng, H.; Li, Y.; Zhang, C.; Wu, S.; Yu, D.; Huang, Z.; Liu, F.; et al. MicroRNA-33b Inhibits Breast Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci. Rep. 2015, 5, 9995. [Google Scholar] [CrossRef]
- Tao, W.Y.; Wang, C.Y.; Sun, Y.H.; Su, Y.H.; Pang, D.; Zhang, G.Q. MicroRNA-34c Suppresses Breast Cancer Migration and Invasion by Targeting GIT1. J. Cancer 2016, 7, 1653. [Google Scholar] [CrossRef] [Green Version]
- Goicoechea, I.; Rezola, R.; Arestin, M.; Caffarel, M.; Cortazar, A.R.; Manterola, L.; Fernandez-Mercado, M.; Armesto, M.; Sole, C.; Larrea, E.; et al. Spatial intratumoural heterogeneity in the expression of GIT1 is associated with poor prognostic outcome in oestrogen receptor positive breast cancer patients with synchronous lymph node metastases. F1000Research 2017, 6, 1606. [Google Scholar] [CrossRef]
- Siemens, H.; Jackstadt, R.; Hunten, S.; Kaller, M.; Menssen, A.; Gotz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Cogdell, D.; Hu, L.; Yang, D.; Sood, A.K.; Xue, F.; Zhang, W. MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol. Rep. 2014, 31, 2021. [Google Scholar] [CrossRef]
- Yue, S.; Wang, L.; Zhang, H.; Min, Y.; Lou, Y.; Sun, H.; Jiang, Y.; Zhang, W.; Liang, A.; Guo, Y.; et al. miR-139-5p suppresses cancer cell migration and invasion through targeting ZEB1 and ZEB2 in GBM. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 6741. [Google Scholar] [CrossRef]
- Hou, Y.; Zhen, J.; Xu, X.; Zhen, K.; Zhu, B.; Pan, R.; Zhao, C. miR-215 functions as a tumor suppressor and directly targets ZEB2 in human non-small cell lung cancer. Oncol. Lett. 2015, 10, 1985. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.B.; Luo, H.P.; Shi, Q.; Hao, Z.N.; Ding, Y.; Wang, Q.S.; Li, S.B.; Xiao, G.C.; Tong, S.L. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J. Gastroenterol. 2014, 20, 6515. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Li, Y.; Fang, N.; Liu, B.; Zu, L.; Chang, R.; Li, X.; Zhou, Q. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS ONE 2014, 9, e91827. [Google Scholar] [CrossRef] [PubMed]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008, 359, 2641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrie, C.H.; Cooper, C.D.; Ballabio, E.; Chi, J.; Tramonti, D.; Hatton, C.S. Aberrant expression of microRNA biosynthetic pathway components is a common feature of haematological malignancy. Br. J. Haematol. 2009, 145, 545. [Google Scholar] [CrossRef] [PubMed]
- Van den Beucken, T.; Koch, E.; Chu, K.; Rupaimoole, R.; Prickaerts, P.; Adriaens, M.; Voncken, J.W.; Harris, A.L.; Buffa, F.M.; Haider, S.; et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat. Commun. 2014, 5, 5203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Chakravarti, D.; Cho, M.S.; Liu, L.; Gi, Y.J.; Lin, Y.L.; Leung, M.L.; El-Naggar, A.; Creighton, C.J.; Suraokar, M.B.; et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010, 467, 986. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Ivan, C.; Yang, D.; Gharpure, K.M.; Wu, S.Y.; Pecot, C.V.; Previs, R.A.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; et al. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene 2016, 35, 4312. [Google Scholar] [CrossRef] [Green Version]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195. [Google Scholar] [CrossRef] [Green Version]
- Garg, M. Emerging role of microRNAs in cancer stem cells: Implications in cancer therapy. World J. Stem Cells 2015, 7, 1078. [Google Scholar] [CrossRef]
- Asadzadeh, Z.; Mansoori, B.; Mohammadi, A.; Aghajani, M.; Haji-Asgarzadeh, K.; Safarzadeh, E.; Mokhtarzadeh, A.; Duijf, P.H.G.; Baradaran, B. microRNAs in cancer stem cells: Biology, pathways, and therapeutic opportunities. J. Cell. Physiol. 2019, 234, 10002. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seton-Rogers, S. Cancer stem cells: Easily moulded. Nat. Rev. Cancer 2013, 13, 519. [Google Scholar] [CrossRef] [PubMed]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef] [Green Version]
- Okuda, H.; Xing, F.; Pandey, P.R.; Sharma, S.; Watabe, M.; Pai, S.K.; Mo, Y.Y.; Iiizumi-Gairani, M.; Hirota, S.; Liu, Y.; et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013, 73, 1434. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858. [Google Scholar] [CrossRef]
- Chu, J.; Li, Y.; Fan, X.; Ma, J.; Li, J.; Lu, G.; Zhang, Y.; Huang, Y.; Li, W.; Huang, X.; et al. MiR-4319 Suppress the Malignancy of Triple-Negative Breast Cancer by Regulating Self-Renewal and Tumorigenesis of Stem Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 48, 593. [Google Scholar] [CrossRef]
- Lv, C.; Li, F.; Li, X.; Tian, Y.; Zhang, Y.; Sheng, X.; Song, Y.; Meng, Q.; Yuan, S.; Luan, L.; et al. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 2017, 8, 1036. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Shin, H.S.; Lee, Y.S.; Lee, Y.C. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab. Investig. A J. Tech. Methods Pathol. 2014, 94, 1370. [Google Scholar] [CrossRef] [PubMed]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of Cellular miRNAs on Circulating miRNA Biomarker Signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Mercado, M.; Manterola, L.; Larrea, E.; Goicoechea, I.; Arestin, M.; Armesto, M.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of non-invasive cancer biomarkers: Concepts and controversies of non-coding and coding RNA in body fluids. J. Cell. Mol. Med. 2015, 19, 2307. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733. [Google Scholar] [CrossRef]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019, 58, 100. [Google Scholar] [CrossRef]
- Aguirre-Ghiso, J.A. On the theory of tumor self-seeding: Implications for metastasis progression in humans. Breast Cancer Res. 2010, 12, 304. [Google Scholar] [CrossRef]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453. [Google Scholar] [CrossRef]
- Aguirre-Ghiso, J.A. How dormant cancer persists and reawakens. Science 2018, 361, 1314. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223. [Google Scholar] [CrossRef]
- Vickers, K.C.; Remaley, A.T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoorvogel, W. Functional transfer of microRNA by exosomes. Blood 2012, 119, 646. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K.; et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, N.; Katsuda, T.; Ochiya, T. Micromanaging of tumor metastasis by extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 52. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Lavotshkin, S.; Lyden, D. The secreted factors responsible for pre-metastatic niche formation: Old sayings and new thoughts. Semin. Cancer Biol. 2011, 21, 139. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lotvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.H.; Zhang, Z.J.; Shang, L.R.; Luo, Y.W.; Lin, Y.F.; Yuan, Y.; Zhuang, S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kgamma to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019, 79, 146. [Google Scholar] [CrossRef] [Green Version]
- Erez, N.; Truitt, M.; Olson, P.; Arron, S.T.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010, 17, 135. [Google Scholar] [CrossRef] [Green Version]
- Josson, S.; Gururajan, M.; Sung, S.Y.; Hu, P.; Shao, C.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q.; et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183. [Google Scholar] [CrossRef] [Green Version]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100. [Google Scholar] [CrossRef]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, P.M.; Loher, P.; Quann, K.; Brody, J.; Londin, E.R.; Rigoutsos, I. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci. Rep. 2014, 4, 5947. [Google Scholar] [CrossRef] [PubMed]
- Ascano, M.; Hafner, M.; Cekan, P.; Gerstberger, S.; Tuschl, T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA 2012, 3, 159. [Google Scholar] [CrossRef] [Green Version]
- Bracken, C.P.; Li, X.; Wright, J.A.; Lawrence, D.M.; Pillman, K.A.; Salmanidis, M.; Anderson, M.A.; Dredge, B.K.; Gregory, P.A.; Tsykin, A.; et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 2014, 33, 2040. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, M.P.; Rajapakshe, K.I.; Bader, D.A.; Cerne, J.Z.; Smith, E.A.; Coarfa, C.; Hartig, S.M.; McGuire, S.E. The Landscape of microRNA Targeting in Prostate Cancer Defined by AGO-PAR-CLIP. Neoplasia 2016, 18, 356. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 2019, 38, 5700. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828. [Google Scholar] [CrossRef]
- Akao, Y.; Nakagawa, Y.; Naoe, T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006, 29, 903. [Google Scholar] [CrossRef] [Green Version]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271. [Google Scholar] [CrossRef]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.P.; Vink, M.A.; Westerink, J.T.; Ramirez de Arellano, E.; Konstantinova, P.; Ter Brake, O.; Berkhout, B. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA 2010, 16, 1328. [Google Scholar] [CrossRef] [Green Version]
- Lou, W.; Chen, Q.; Ma, L.; Liu, J.; Yang, Z.; Shen, J.; Cui, Y.; Bian, X.W.; Qian, C. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J. Mol. Med. 2013, 91, 715. [Google Scholar] [CrossRef]
- Collins, S.A.; Guinn, B.A.; Harrison, P.T.; Scallan, M.F.; O’Sullivan, G.C.; Tangney, M. Viral vectors in cancer immunotherapy: Which vector for which strategy? Curr. Gene Ther. 2008, 8, 66. [Google Scholar] [CrossRef]
- Campani, V.; Salzano, G.; Lusa, S.; De Rosa, G. Lipid Nanovectors to Deliver RNA Oligonucleotides in Cancer. Nanomaterials 2016, 6, 131. [Google Scholar] [CrossRef] [Green Version]
- Ando, H.; Okamoto, A.; Yokota, M.; Asai, T.; Dewa, T.; Oku, N. Polycation liposomes as a vector for potential intracellular delivery of microRNA. J. Gene Med. 2013, 15, 375. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, X. MicroRNA Delivery with Bioreducible Polyethylenimine as a Non-Viral Vector for Breast Cancer Gene Therapy. Macromol. Biosci. 2019, 19, e1800445. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, Y.; Zhang, X.; Chen, J. Three-layered polyplex as a microRNA targeted delivery system for breast cancer gene therapy. Nanotechnology 2017, 28, 285101. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Q.; Yuan, Y.; Gong, Y.; Luo, X.; Su, X.; Hu, X.; Zhu, W. Therapeutic delivery of microRNA-143 by cationic lipoplexes for non-small cell lung cancer treatment in vivo. J. Cancer Res. Clin. Oncol. 2019, 145, 2951. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Yi, B.; Ma, R.; Zhang, X.; Zhao, H.; Xi, Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016, 6, 22312. [Google Scholar] [CrossRef] [Green Version]
- Aquino-Jarquin, G. Emerging Role of CRISPR/Cas9 Technology for MicroRNAs Editing in Cancer Research. Cancer Res. 2017, 77, 6812. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Meng, X.; Pan, J.; Jiang, N.; Zhou, C.; Wu, Z.; Gong, Z. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 2018, 15, 35. [Google Scholar] [CrossRef] [Green Version]
- Huo, W.; Zhao, G.; Yin, J.; Ouyang, X.; Wang, Y.; Yang, C.; Wang, B.; Dong, P.; Wang, Z.; Watari, H.; et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J. Cancer 2017, 8, 57. [Google Scholar] [CrossRef]
- El Fatimy, R.; Subramanian, S.; Uhlmann, E.J.; Krichevsky, A.M. Genome Editing Reveals Glioblastoma Addiction to MicroRNA-10b. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 368. [Google Scholar] [CrossRef] [Green Version]
- Esposito, C.L.; Cerchia, L.; Catuogno, S.; De Vita, G.; Dassie, J.P.; Santamaria, G.; Swiderski, P.; Condorelli, G.; Giangrande, P.H.; de Franciscis, V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumor microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almanza, G.; Rodvold, J.J.; Tsui, B.; Jepsen, K.; Carter, H.; Zanetti, M. Extracellular vesicles produced in B cells deliver tumor suppressor miR-335 to breast cancer cells disrupting oncogenic programming in vitro and in vivo. Sci. Rep. 2018, 8, 17581. [Google Scholar] [CrossRef] [PubMed]
- Van Roosbroeck, K.; Fanini, F.; Setoyama, T.; Ivan, C.; Rodriguez-Aguayo, C.; Fuentes-Mattei, E.; Xiao, L.; Vannini, I.; Redis, R.S.; D’Abundo, L.; et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 2017, 23, 2891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, M.; Sekar, T.V.; Ananta, J.S.; Devulapally, R.; Afjei, R.; Babikir, H.A.; Paulmurugan, R.; Massoud, T.F. Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of temozolomide in vitro and in a mouse model. Oncotarget 2018, 9, 21478. [Google Scholar] [CrossRef] [Green Version]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 2015, 9, 2290. [Google Scholar] [CrossRef] [Green Version]
- Cortez, M.A.; Valdecanas, D.; Zhang, X.; Zhan, Y.; Bhardwaj, V.; Calin, G.A.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1494. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.T.; Pongkulapa, T.; Cho, H.Y.; Han, J.; Pasquale, N.J.; Rabie, H.; Kim, J.H.; Choi, J.W.; Lee, K.B. Overcoming Chemoresistance in Cancer via Combined MicroRNA Therapeutics with Anticancer Drugs Using Multifunctional Magnetic Core-Shell Nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 26954. [Google Scholar] [CrossRef]
- Rui, M.; Qu, Y.; Gao, T.; Ge, Y.; Feng, C.; Xu, X. Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int. J. Nanomed. 2017, 12, 217. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhang, L.; Bai, X.; Cao, X.; Jiao, X.; Huang, Y.; Li, Y.; Qin, Y.; Wen, Y. Stimuli-Responsive Nanocarrier for Co-delivery of MiR-31 and Doxorubicin To Suppress High MtEF4 Cancer. ACS Appl. Mater. Interfaces 2018, 10, 22767. [Google Scholar] [CrossRef]
- Yao, C.; Liu, J.; Wu, X.; Tai, Z.; Gao, Y.; Zhu, Q.; Li, J.; Zhang, L.; Hu, C.; Gu, F.; et al. Reducible self-assembling cationic polypeptide-based micelles mediate co-delivery of doxorubicin and microRNA-34a for androgen-independent prostate cancer therapy. J. Control. Release 2016, 232, 203. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Fan, W.; Wang, Y.; Huang, L.; Jiang, Y.; Shi, L.; McKinley, D.; Tan, W.; Tan, C. Combined Delivery of Let-7b MicroRNA and Paclitaxel via Biodegradable Nanoassemblies for the Treatment of KRAS Mutant Cancer. Mol. Pharm. 2016, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Uz, M.; Kalaga, M.; Pothuraju, R.; Ju, J.; Junker, W.M.; Batra, S.K.; Mallapragada, S.; Rachagani, S. Dual delivery nanoscale device for miR-345 and gemcitabine co-delivery to treat pancreatic cancer. J. Control. Release 2019, 294, 237. [Google Scholar] [CrossRef] [PubMed]
- Mondal, G.; Almawash, S.; Chaudhary, A.K.; Mahato, R.I. EGFR-Targeted Cationic Polymeric Mixed Micelles for Codelivery of Gemcitabine and miR-205 for Treating Advanced Pancreatic Cancer. Mol. Pharm. 2017, 14, 3121. [Google Scholar] [CrossRef]
- Xu, J.; Sun, J.; Ho, P.Y.; Luo, Z.; Ma, W.; Zhao, W.; Rathod, S.B.; Fernandez, C.A.; Venkataramanan, R.; Xie, W.; et al. Creatine based polymer for codelivery of bioengineered MicroRNA and chemodrugs against breast cancer lung metastasis. Biomaterials 2019, 210, 25. [Google Scholar] [CrossRef]
- Bader, A.G. miR-34—A microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, 120. [Google Scholar] [CrossRef] [Green Version]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079. [Google Scholar] [CrossRef] [Green Version]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386. [Google Scholar] [CrossRef]
- Haghi, M.; Taha, M.F.; Javeri, A. Suppressive effect of exogenous miR-16 and miR-34a on tumorigenesis of breast cancer cells. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Xu, M.; Li, D.; Yang, C.; Ji, J.S. MicroRNA-34a Inhibition of the TLR Signaling Pathway Via CXCL10 Suppresses Breast Cancer Cell Invasion and Migration. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 1286. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.H.; Yeh, C.T.; Ho, J.Y.; Ng, K.F.; Chen, T.C. OncomiR miR-96 and miR-182 promote cell proliferation and invasion through targeting ephrinA5 in hepatocellular carcinoma. Mol. Carcinog. 2016, 55, 366. [Google Scholar] [CrossRef]
- Han, L.; Chen, W.; Xia, Y.; Song, Y.; Zhao, Z.; Cheng, H.; Jiang, T. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am. J. Transl. Res. 2018, 10, 1172. [Google Scholar]
- Ji, H.; Sang, M.; Liu, F.; Ai, N.; Geng, C. miR-124 regulates EMT based on ZEB2 target to inhibit invasion and metastasis in triple-negative breast cancer. Pathol. Res. Pract. 2019, 215, 697. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, R.; Xu, S.; Li, Y.; Zhao, P.; Dong, W.; Liu, Z.; Zhao, Q.; Tan, B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim. Et Biophys. Sin. 2018, 50, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Du, R.; Zhou, L.; Xu, J.; Chen, S.; Chen, J.; Yang, X.; Liu, D.X.; Shao, Z.M.; Zhang, L.; et al. miR-200c/141 Regulates Breast Cancer Stem Cell Heterogeneity via Targeting HIPK1/beta-Catenin Axis. Theranostics 2018, 8, 5801. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xia, L.; Zhou, Z.; Zuo, Z.; Xu, C.; Song, H.; Cai, J. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch. Biochem. Biophys. 2018, 640, 53. [Google Scholar] [CrossRef]
- Sun, G.; Liu, M.; Han, H. Overexpression of microRNA-190 inhibits migration, invasion, epithelial-mesenchymal transition, and angiogenesis through suppression of protein kinase B-extracellular signal-regulated kinase signaling pathway via binding to stanniocalicin 2 in breast cancer. J. Cell. Physiol. 2019, 10, 17824. [Google Scholar]
- Yu, Y.; Luo, W.; Yang, Z.J.; Chi, J.R.; Li, Y.R.; Ding, Y.; Ge, J.; Wang, X.; Cao, X.C. miR-190 suppresses breast cancer metastasis by regulation of TGF-beta-induced epithelial-mesenchymal transition. Mol. Cancer 2018, 17, 70. [Google Scholar] [CrossRef] [Green Version]
- Bian, X.; Liang, Z.; Feng, A.; Salgado, E.; Shim, H. HDAC inhibitor suppresses proliferation and invasion of breast cancer cells through regulation of miR-200c targeting CRKL. Biochem. Pharmacol. 2018, 147, 30. [Google Scholar] [CrossRef]
- Liu, J.; He, D.; Zhang, G.J. Bioluminescence Imaging for Monitoring miR-200c Expression in Breast Cancer Cells and its Effects on Epithelial-Mesenchymal Transition Progress in Living Animals. Mol. Imaging Biol. Mib Off. Publ. Acad. Mol. Imaging 2018, 20, 761. [Google Scholar] [CrossRef]
- Seo, S.; Moon, Y.; Choi, J.; Yoon, S.; Jung, K.H.; Cheon, J.; Kim, W.; Kim, D.; Lee, C.H.; Kim, S.W.; et al. The GTP binding activity of transglutaminase 2 promotes bone metastasis of breast cancer cells by downregulating microRNA-205. Am. J. Cancer Res. 2019, 9, 597. [Google Scholar] [PubMed]
- Wu, L.; Zhang, Y.; Huang, Z.; Gu, H.; Zhou, K.; Yin, X.; Xu, J. MiR-409-3p Inhibits Cell Proliferation and Invasion of Osteosarcoma by Targeting Zinc-Finger E-Box-Binding Homeobox-1. Front. Pharmacol. 2019, 10, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.J.; Zeng, H.X.; Huang, P.; Wang, S.; Xie, C.H.; Li, S.J. MiR-508-3p inhibits cell invasion and epithelial-mesenchymal transition by targeting ZEB1 in triple-negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6379. [Google Scholar] [PubMed]
- Yao, R.; Zheng, H.; Wu, L.; Cai, P. miRNA-641 inhibits the proliferation, migration, and invasion and induces apoptosis of cervical cancer cells by directly targeting ZEB1. Onco Targets Ther. 2018, 11, 8965. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.W.; Guan, W.; Han, S.; Hong, D.K.; Kim, L.S.; Kim, H. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018, 109, 1404. [Google Scholar] [CrossRef] [Green Version]


| miRNA | Cancer | Express. | Sample | Target | Ref |
|---|---|---|---|---|---|
| Let-7 | Breast | Low | Cells & Tissue | RAS & HMGA2 | [14] |
| miR-7 | Breast | Low | Cells & Tissue | FAK | [58] |
| miR-10b | Breast | High | Cells & Tissue | HOXD10 | [13] |
| miR-26a/b | HNSCC | Low | Cells & Tissue | LOXL2 | [45] |
| miR-29 family | HNSCC | Low | Cells | LAMC2 & ITGA6 | [44] |
| miR-29 family | HNSCC | Low | Cells & Tissue | LOXL2 | [45] |
| miR-29 family | ccRCC | Low | Cells & Tissue | LOXL2 | [46] |
| miR-29a | Breast | High | Cells & Tissue | SUV420H2 | [25] |
| miR-29c | Lung | Low | Cells | Integrin β1 & MMP2 | [48] |
| miR-31 | Breast | Low | Cells | Integrin α subunits | [60] |
| miR-31 | Breast | Low | Cells & Tissue | WAVE3 | [59] |
| miR-33b | Breast | Low | Cells & Tissue | HMGA2, SALL4 & Twist1 | [65] |
| miR-34 | CRC | Low | Cells | ZEB1 & Slug | [68] |
| miR-34a/b/c | CRC | Low | Cells | Snail | [170] |
| miR-34a | Breast | Low | Cells & Tissue | CXCL10 | [171] |
| miR-34c | Breast | Low | Cells & Tissue | GIT1 | [66] |
| miR-93 | Endometrial | High | Cells & Tissue | FOXA1 | [29] |
| miR-96 | Breast | High | Cells & Tissue | PTPN9 | [23] |
| miR-96 | HCC | High | Cells & Tissue | ephrinA5 | [172] |
| miR-101 | Ovarian | Low | Cells | ZEB1 & ZEB2 | [69] |
| miR-101 | NSCLC | Low | Cells & Tissue | ZEB1 | [173] |
| miR-105 | Breast | High | Cells, Tissue & Serum | ZO-1 | [17] |
| miR-124 | Breast | Low | Cells & Tissue | ZEB2 | [174] |
| miR-124 | Breast | Low | Cells & Tissue | IL-11 | [63] |
| miR-124 | CRC | Low | Cells & Tissue | ROCK1 | [55] |
| miR-128-3p | ESCC | Low | Cells & Tissue | ZEB1 | [175] |
| miR-130a | Breast | Low | Cells | FOSL1 | [26] |
| miR-132 | NSCLC | Low | Cells & Tissue | ZEB2 | [73] |
| miR-132 | CRC | Low | Cells & Tissue | ZEB2 | [72] |
| miR-135a | Breast | High | Cells & Tissue | HOXA10 | [20] |
| miR-135a | HCC | High | Cells & Tissue | FOXO1 | [22] |
| miR-135a | Gastric | Low | Cells & Tissue | ROCK1 | [56] |
| miR-135b | Breast | High | Cells & Tissue | APC | [21] |
| miR-138 | Breast | Low | Cells & Tissue | ROCK1 | [57] |
| miR-139-5p | Glioblastoma | Low | Cells & Tissue | ZEB1 & ZEB2 | [70] |
| miR-141 | Breast | Low | Cells & Tissue | HIPK1 | [176] |
| miR-141 | Prostate | Low | Cells & Tissue | Rho GTases, CD44 & EZH2 | [43] |
| miR-150 | Breast | Low | Cells & Tissue | HMGA2 | [62] |
| miR-154 | Breast | Low | Cells & Tissue | E2F5 | [61] |
| miR-181b | Breast | High | Cells & Tissue | YWHAG | [18] |
| miR-182 | HCC | High | Cells & Tissue | ephrinA5 | [172] |
| miR-186-5p | CRC | Low | Cells | ZEB1 | [177] |
| miR-190 | Breast | Low | Cells | STC2 | [178] |
| miR-190 | Breast | Low | Cells & Tissue | SMAD2 | [179] |
| miR-194 | Ovarian | High | Cells & Tissue | PTPN12 | [37] |
| miR-197 | HCC | High | Cells & Tissue | Axin-2, NKD1 & DKK2 | [30] |
| miR-200 | Breast | Low | Cells & Tissue | ZEB1 & ZEB2 | [39] |
| miR-200a/b | Breast | Low | Cells | ZEB1 & ZEB2 | [38] |
| miR-200c | Breast | Low | Cells | CRKL | [180] |
| miR-200c | Breast | Low | Cells | ZEB1 | [181] |
| miR-200c | Breast | Low | Cells & Tissue | HIPK1 | [176] |
| miR-203 | HNSCC | Low | Cells & Tissue | LASP1, SPARC & NUAK | [49] |
| miR-203 | Breast | Low | Cells | Slug | [50] |
| miR-203 | CRC | Low | Cells & Tissue | EIF5A2 | [54] |
| miR-203 | Melanoma | Low | Cells & Tissue | Slug | [51] |
| miR-203 | Ovarian | Low | Cells & Tissue | BIRC5 | [52] |
| miR-203 | Gastric | Low | Cells & Tissue | Annexin A4 | [53] |
| miR-205 | Breast | Low | Cells | ZEB1 | [182] |
| miR-205 | Breast | Low | Cells & Tissue | ZEB1 & ZEB2 | [39] |
| miR-210 | Breast | High | Cells & Tissue | E-Cadherin & Snail | [24] |
| miR-215 | NSCLC | Low | Cells & Tissue | ZEB2 | [71] |
| miR-218 | HNSCC | Low | Cells & Tissue | LOXL2 | [45] |
| miR-374a | Breast | High | Cells & Tissue | WIF1, PTEN & WNT5A | [19] |
| miR-409-3p | Osteosarcoma | Low | Cells & Tissue | ZEB1 | [183] |
| miR-429 | Breast | Low | Cells | ZEB1 & ZEB2 | [38] |
| miR-504 | NSCLC | Low | Cells & Tissue | LOXL2 | [47] |
| miR-508-3p | Breast | Low | Cells & Tissue | ZEB1 | [184] |
| miR-520c | Gastic | High | Cells & Tissue | IRF2 | [32] |
| miR-574-5p | NSCLC | High | Cells, Tissue & Serum | PTPRU | [36] |
| miR-577 | Gastric | High | Cells & Tissue | SDPR | [31] |
| miR-641 | Cervical | Low | Cells & Tissue | ZEB1 | [185] |
| miR-708-3p | Breast | Low | Cells & Tissue | ZEB1, CDH2 & Vimentin | [186] |
| miR-1204 | Breast | High | Cells & Tissue | VDR | [28] |
| miR-1269a | Colorectal | High | Cells & Tissue | Smad7 & HOXD10 | [33] |
| miR-8084 | Breast | High | Cells, Tissue & Serum | ING2 | [27] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solé, C.; Lawrie, C.H. MicroRNAs and Metastasis. Cancers 2020, 12, 96. https://doi.org/10.3390/cancers12010096
Solé C, Lawrie CH. MicroRNAs and Metastasis. Cancers. 2020; 12(1):96. https://doi.org/10.3390/cancers12010096
Chicago/Turabian StyleSolé, Carla, and Charles H. Lawrie. 2020. "MicroRNAs and Metastasis" Cancers 12, no. 1: 96. https://doi.org/10.3390/cancers12010096




