Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme
Abstract
:1. Introduction
2. Results
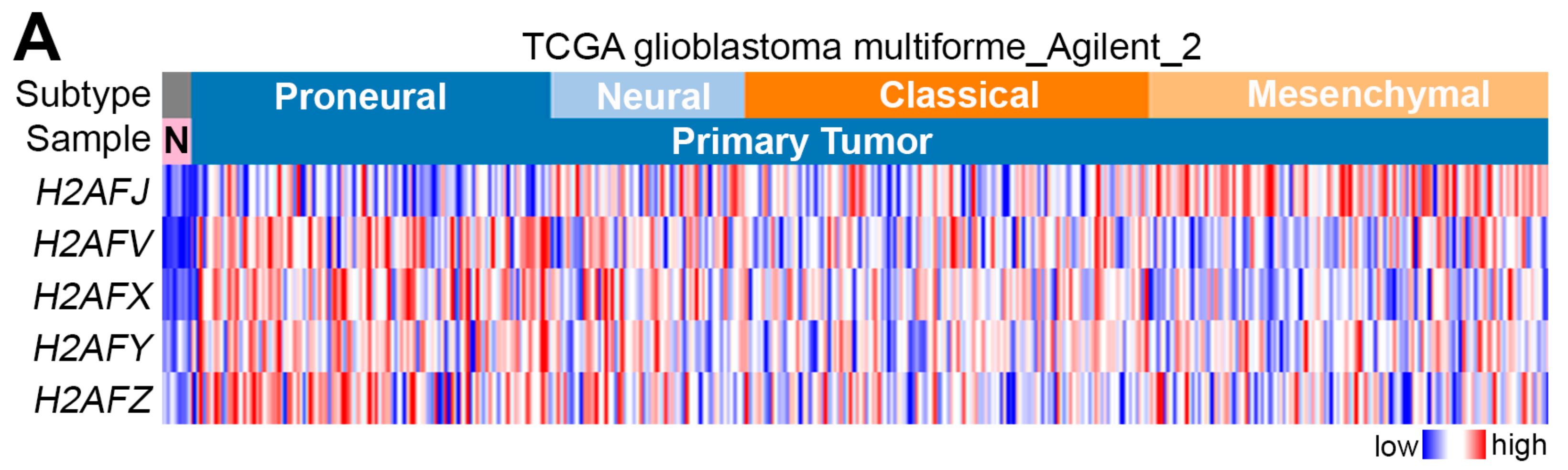
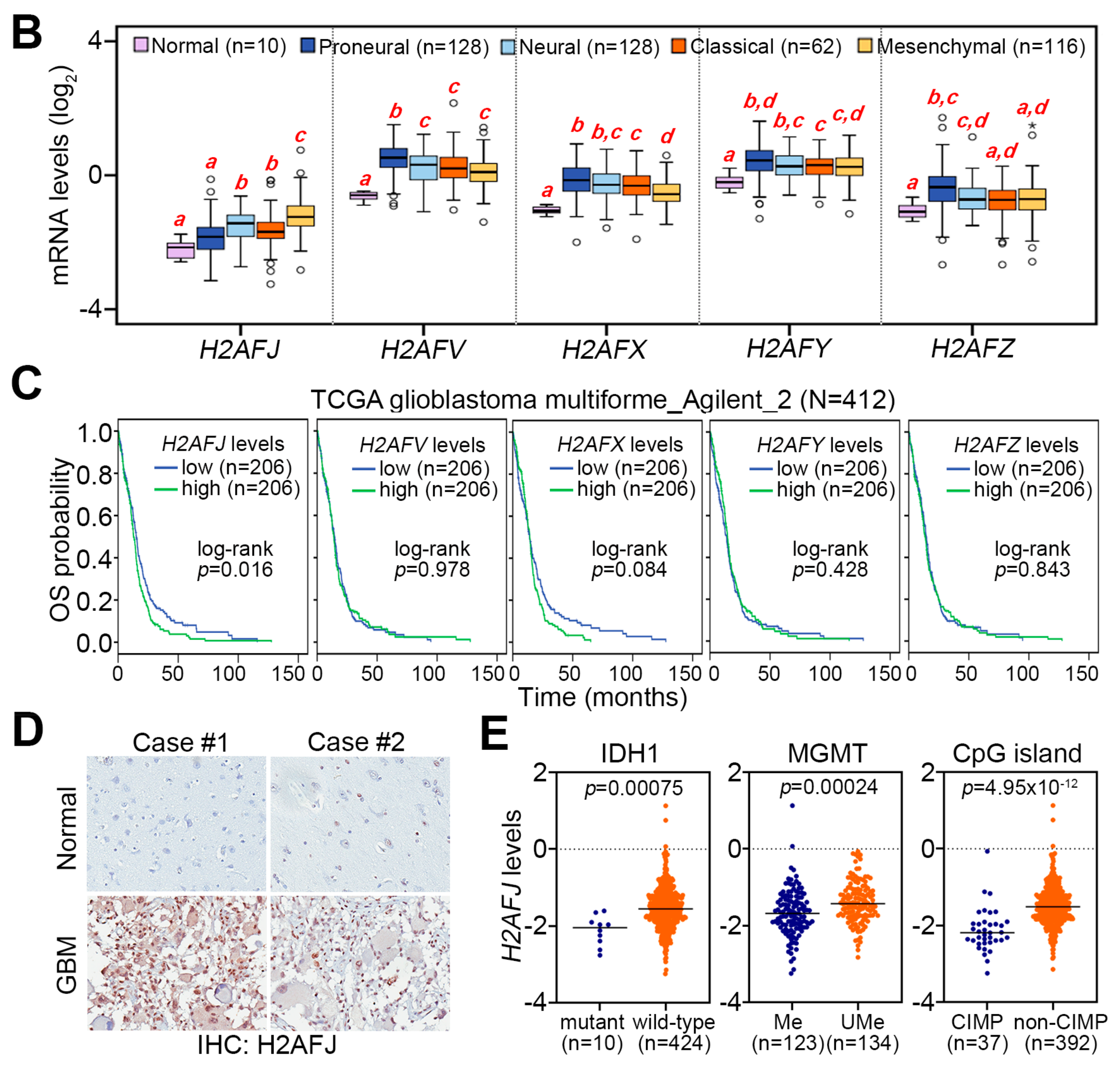
2.1. H2AFJ Is Frequently Upregulated in Mesenchymal-Type GBM Compared to Normal Brain Tissues and Low-Grade Gliomas
2.2. H2AFJ Upregulation Is Highly Correlated with a Poor Prognosis in Patients with Brain Tumors
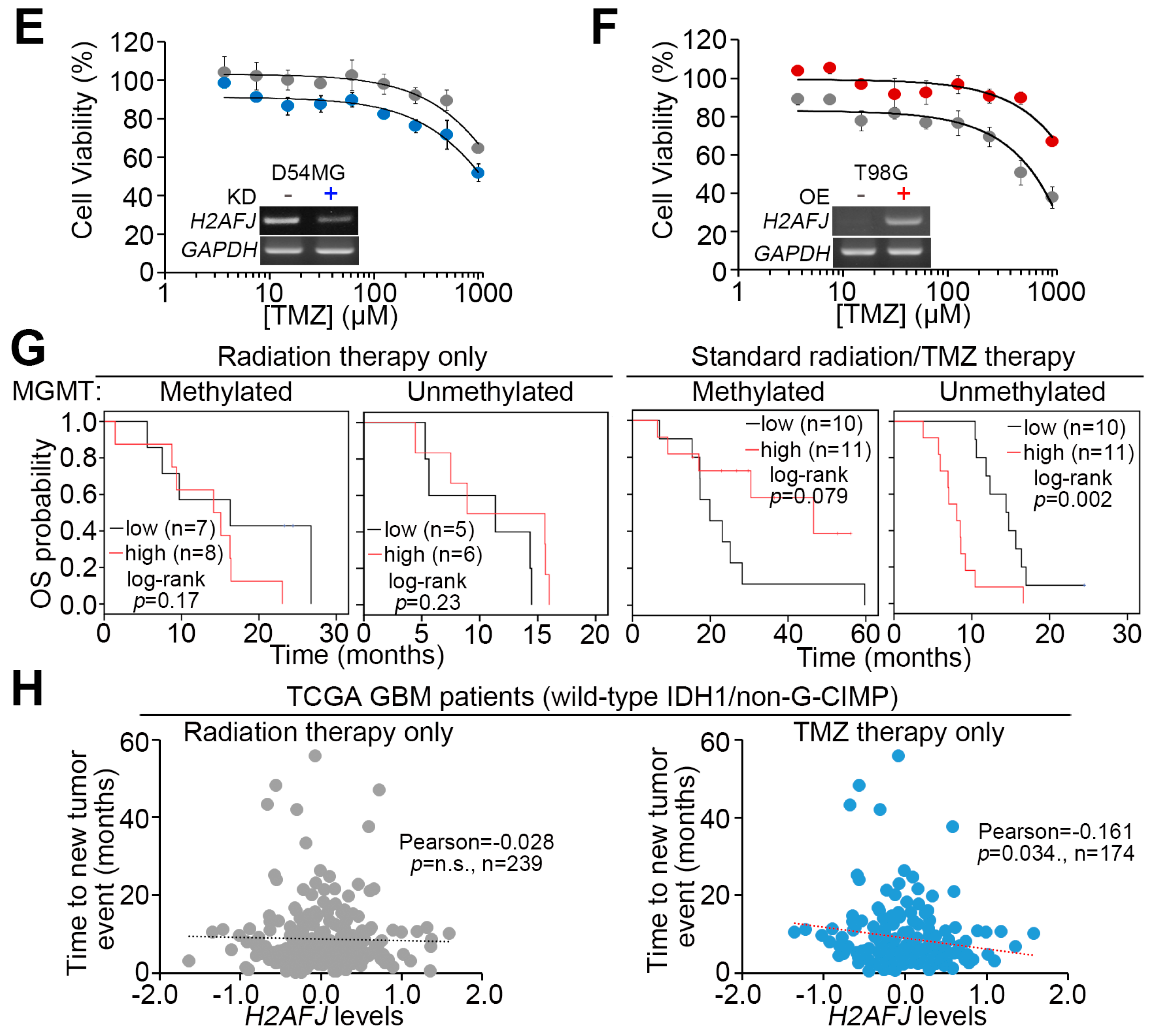
2.3. H2AFJ Expression Determines TMZ, not Radiation, Effectiveness in GBM.
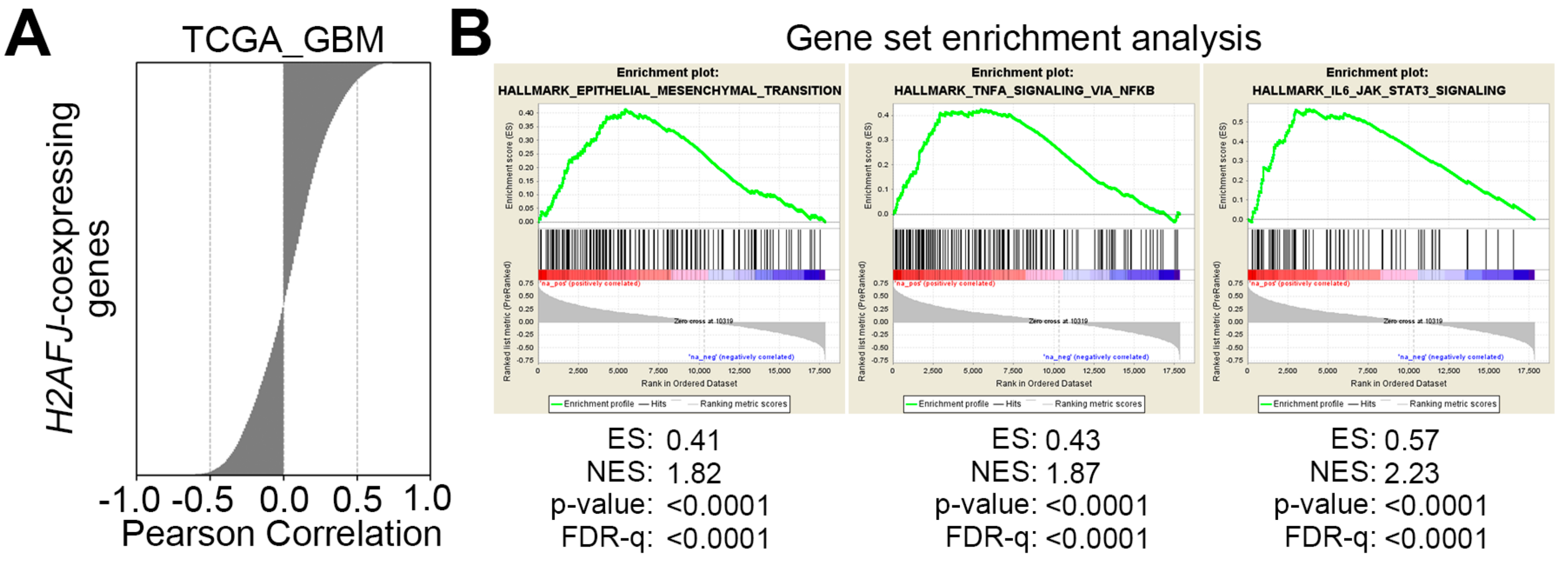
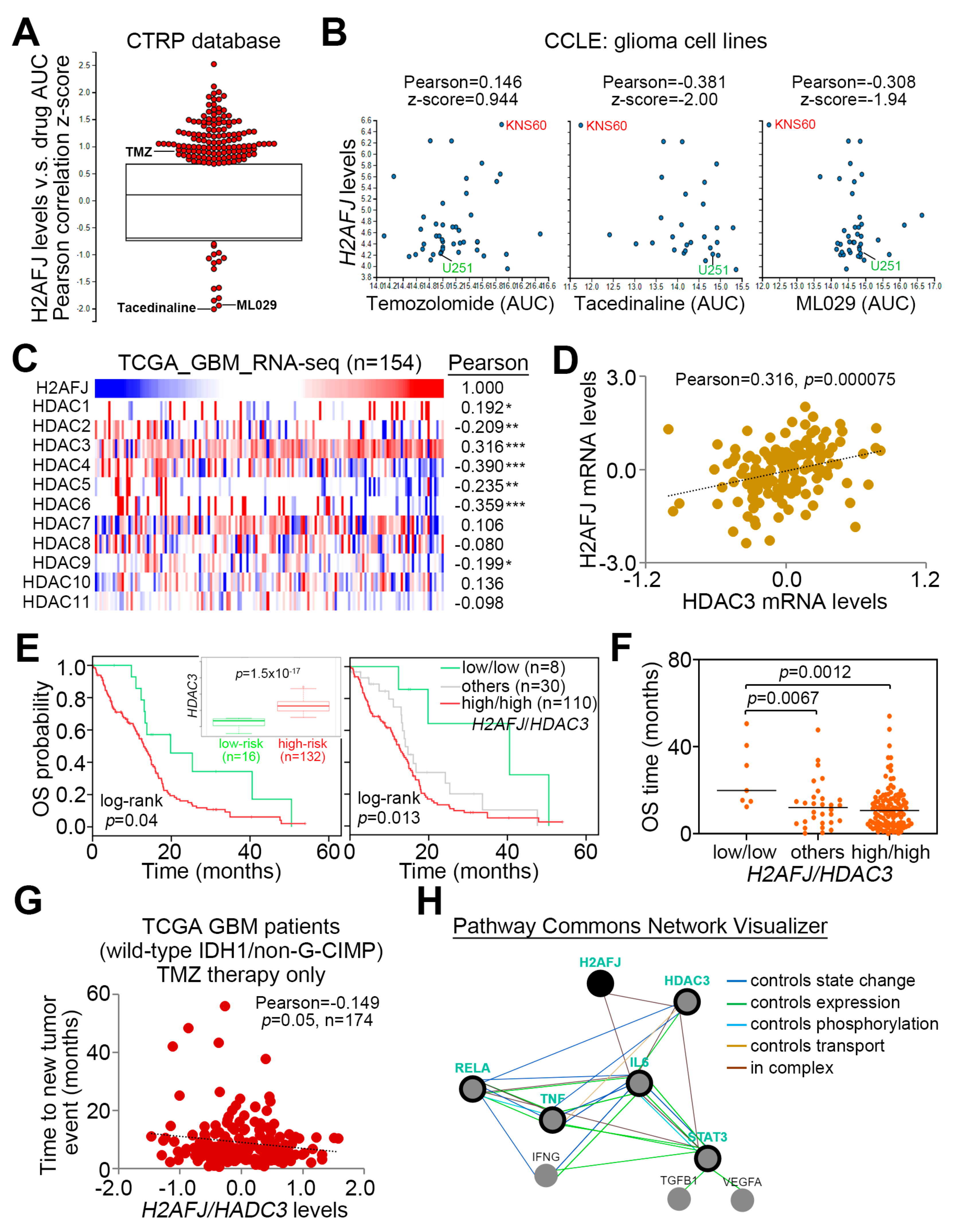
2.4. H2AFJ Upregulation Associates with the Progression of Proneural-Mesenchymal Transition and the Activation of TNFα-NF-κB and IL6-STAT3 Signaling Pathways in GBM
2.5. Pharmaceutical Inhibition of NF-κB and HDAC3 Provides a New Strategy for Overcoming TMZ-Insensitive GBM with H2AFJ Upregulation
3. Discussion
4. Materials and Methods
4.1. Clinical and Molecular Data for Patients
4.2. Cell Lines and Cell Culture Conditions
4.3. MTT Assay
4.4. Plasmid Construction
4.5. Lentivirus-Driven shRNA Infection
4.6. Reverse Transcription PCR (RT-PCR)
4.7. Irradiation Treatment and Cell Viability Analysis
4.8. Luciferase Reporter Assay
4.9. Immunohistochemistry Staining Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von, D.A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandana, S.R.; Movva, S.; Arora, M.; Singh, T. Primary brain tumors in adults. Am. Fam. Physician 2008, 77, 1423–1430. [Google Scholar] [PubMed]
- Urbanska, K.; Sokolowska, J.; Szmidt, M.; Sysa, P. Glioblastoma multiforme—An overview. Contemp. Oncol. (Pozn.) 2014, 18, 307–312. [Google Scholar] [PubMed]
- Ohka, F.; Natsume, A.; Wakabayashi, T. Current trends in targeted therapies for glioblastoma multiforme. Neurol. Res. Int. 2012, 2012, 878425. [Google Scholar] [CrossRef]
- Batash, R.; Asna, N.; Schaffer, P.; Francis, N.; Schaffer, M. Glioblastoma Multiforme, Diagnosis and Treatment; Recent Literature Review. Curr. Med. Chem. 2017, 24, 3002–3009. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Moore, L.M.; Li, X.; Yung, W.K.; Zhang, W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol. 2013, 15, 1114–1126. [Google Scholar] [CrossRef]
- Labussiere, M.; Idbaih, A.; Wang, X.W.; Marie, Y.; Boisselier, B.; Falet, C.; Paris, S.; Laffaire, J.; Carpentier, C.; Criniere, E.; et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology 2010, 74, 1886–1890. [Google Scholar] [CrossRef]
- Dubbink, H.J.; Taal, W.; Van Marion, R.; Kros, J.M.; Van Heuvel, I.; Bromberg, J.E.; Zonnenberg, B.A.; Zonnenberg, C.; Postma, T.J.; Gijtenbeek, J.M.; et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurol 2009, 73, 1792–1795. [Google Scholar] [CrossRef]
- Minniti, G.; Scaringi, C.; Arcella, A.; Lanzetta, G.; Di, S.D.; Scarpino, S.; Bozzao, A.; Pace, A.; Villani, V.; Salvati, M.; et al. IDH1 mutation and MGMT methylation status predict survival in patients with anaplastic astrocytoma treated with temozolomide-based chemoradiotherapy. J. Neurooncol. 2014, 118, 377–383. [Google Scholar] [CrossRef]
- Brandes, A.A.; Franceschi, E.; Tosoni, A.; Blatt, V.; Pession, A.; Tallini, G.; Bertorelle, R.; Bartolini, S.; Calbucci, F.; Andreoli, A.; et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J. Clin. Oncol. 2008, 26, 2192–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, J.; Cheng, G.; Zhang, J.; Li, X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin. Neurol. Neurosurg. 2016, 151, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Malakhov, N.; Sheth, N.; Wang, A.; Han, P.; Schreiber, D. Patterns of care and outcomes of chemoradiation versus radiation alone for MGMT promoter unmethylated glioblastoma. Clin. Neurol. Neurosurg. 2018, 170, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Jasencakova, Z.; Groth, A. Chromatin Replication and Histone Dynamics. Adv. Exp. Med. Biol. 2017, 1042, 311–333. [Google Scholar] [PubMed]
- Testa, E.; Nardozi, D.; Antinozzi, C.; Faieta, M.; Di Cecca, S.; Caggiano, C.; Fukuda, T.; Bonanno, E.; Zhenkun, L.; Maldonado, A.; et al. H2AFX and MDC1 promote maintenance of genomic integrity in male germ cells. J. Cell Sci. 2018, 131, jcs214411. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, F.L.; Vitorino, R.; Wang, J.; Cardoso, H.; Laranjeira, H.; Simoes, J.; Caldas, M.; Henrique, R.; Amado, F.; Williams, C.; et al. The histone H2A isoform Hist2h2ac is a novel regulator of proliferation and epithelial-mesenchymal transition in mammary epithelial and in breast cancer cells. Cancer Lett. 2017, 396, 42–52. [Google Scholar] [CrossRef]
- Fan, W.; Zhou, K.; Zhao, Y.; Wu, W.; Chen, H.; Jin, L.; Chen, G.; Shi, J.; Wei, Q.; Zhang, T.; et al. Possible association between genetic variants in the H2AFX promoter region and risk of adult glioma in a Chinese Han population. J. Neurooncol. 2011, 105, 211–218. [Google Scholar] [CrossRef]
- Juarez-Velazquez, R.; Reyes-Leon, A.; Salas-Labadia, C.; Rivera-Luna, R.; Velasco-Hidalgo, L.; Lopez-Hernandez, G.; Lopez-Santiago, N.; Paredes-Aguilera, R.; Dominguez-Lopez, A.; Bernaldez, R.; et al. Significance of CASP8AP2 and H2AFZ expression in survival and risk of relapse in children with acute lymphoblastic leukemia. Leuk. Lymphoma 2014, 55, 2305–2311. [Google Scholar] [CrossRef]
- Jin, X.M.; Kim, H.N.; Shin, M.H.; Lee, I.K.; Lee, J.S.; Lee, J.H.; Kim, H.J.; Choi, J.S.; Juhng, S.W.; Choi, C. H2AFX polymorphisms are associated with decreased risk of diffuse large B cell lymphoma in Koreans. DNA Cell Biol. 2011, 30, 1039–1044. [Google Scholar] [CrossRef]
- Segerman, A.; Niklasson, M.; Haglund, C.; Bergstrom, T.; Jarvius, M.; Xie, Y.; Westermark, A.; Sonmez, D.; Hermansson, A.; Kastemar, M.; et al. Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Transition. Cell Rep. 2016, 17, 2994–3009. [Google Scholar] [CrossRef] [Green Version]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, D.A.; Boehm, K.A.; Waterhouse, D.M.; Wagener, D.J.; Krishnamurthi, S.S.; Rosemurgy, A.; Grove, W.; Macdonald, K.; Gulyas, S.; Clark, M.; et al. Gemcitabine plus CI-994 offers no advantage over gemcitabine alone in the treatment of patients with advanced pancreatic cancer: Results of a phase II randomized, double-blind, placebo-controlled, multicenter study. Ann. Oncol. 2006, 17, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martinez-Ledesma, E.; Martinez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Trevino, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.F.; Mao, X.Y.; Zhu, T.; Mao, C.X.; Liu, Z.X.; Wang, Z.B.; Li, L.; Li, X.; Yin, J.Y.; Zhang, W.; et al. COL3A1 and SNAP91: Novel glioblastoma markers with diagnostic and prognostic value. Oncotarget 2016, 7, 70494–70503. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, S.; Pu, J.K.; Tsang, A.C.; Lee, D.; Man, V.O.; Lui, W.M.; Wong, S.T.; Leung, G.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef]
- Seashore-Ludlow, B.; Rees, M.G.; Cheah, J.H.; Cokol, M.; Price, E.V.; Coletti, M.E.; Jones, V.; Bodycombe, N.E.; Soule, C.K.; Gould, J.; et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015, 5, 1210–1223. [Google Scholar] [CrossRef] [Green Version]
- Olmez, I.; Love, S.; Xiao, A.; Manigat, L.; Randolph, P.; McKenna, B.D.; Neal, B.P.; Boroda, S.; Li, M.; Brenneman, B.; et al. Targeting the mesenchymal subtype in glioblastoma and other cancers via inhibition of diacylglycerol kinase alpha. Neuro Oncol. 2018, 20, 192–202. [Google Scholar] [CrossRef] [Green Version]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de, T.N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Chen, Y.; Wang, S.; Li, P.; Zhou, G.; Yuan, Y. Inhibition of NF-kappaB results in anti-glioma activity and reduces temozolomide-induced chemoresistance by down-regulating MGMT gene expression. Cancer Lett. 2018, 428, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Brassesco, M.S.; Roberto, G.M.; Morales, A.G.; Oliveira, J.C.; Delsin, L.E.; Pezuk, J.A.; Valera, E.T.; Carlotti, C.G., Jr.; Rego, E.M.; de Oliveira, H.F.; et al. Inhibition of NF-kappa B by Dehydroxymethylepoxyquinomicin Suppresses Invasion and Synergistically Potentiates Temozolomide and gamma -Radiation Cytotoxicity in Glioblastoma Cells. Chemother. Res. Pract. 2013, 2013, 593020. [Google Scholar] [PubMed] [Green Version]
- Bredel, M.; Bredel, C.; Juric, D.; Duran, G.E.; Yu, R.X.; Harsh, G.R.; Vogel, H.; Recht, L.D.; Scheck, A.C.; Sikic, B.I. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J. Clin. Oncol. 2006, 24, 274–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, C.; Penault-Llorca, F.; Kemeny, J.L.; Lemaire, J.J.; Jullien, C.; Labit-Bouvier, C.; Finat-Duclos, F.; Verrelle, P. Interleukin-6 overexpression as a marker of malignancy in human gliomas. J. Neurosurg. 2001, 94, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Vlachostergios, P.J.; Hatzidaki, E.; Papandreou, C.N. MGMT repletion after treatment of glioblastoma cells with temozolomide and O6-benzylguanine implicates NFkappaB and mutant p53. Neurol. Res. 2013, 35, 879–882. [Google Scholar] [CrossRef]
- Bredel, M.; Scholtens, D.M.; Yadav, A.K.; Alvarez, A.A.; Renfrow, J.J.; Chandler, J.P.; Yu, I.L.; Carro, M.S.; Dai, F.; Tagge, M.J.; et al. NFKBIA deletion in glioblastomas. N. Engl. J. Med. 2011, 364, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Song, N.; Liu, J.; An, S.; Nishino, T.; Hishikawa, Y.; Koji, T. Immunohistochemical Analysis of Histone H3 Modifications in Germ Cells during Mouse Spermatogenesis. Acta Histochem. Cytochem. 2011, 44, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [Green Version]
- McCabe, M.T.; Mohammad, H.P.; Barbash, O.; Kruger, R.G. Targeting Histone Methylation in Cancer. Cancer J. 2017, 23, 292–301. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Waterborg, J.H. Evolution of histone H3: Emergence of variants and conservation of post-translational modification sites. Biochem. Cell Biol. 2012, 90, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Corujo, D.; Buschbeck, M. Post-Translational Modifications of H2A Histone Variants and Their Role in Cancer. Cancers 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthel, F.P.; Johnson, K.C.; Varn, F.S.; Moskalik, A.D.; Tanner, G.; Kocakavuk, E.; Anderson, K.J.; Abiola, O.; Aldape, K.; Alfaro, K.D.; et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019, 576, 112–120. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-H.; Lin, C.-H.; Lin, H.-Y.; Kuei, C.-H.; Zheng, J.-Q.; Wang, Y.-H.; Lu, L.-S.; Lee, F.-P.; Hu, C.-J.; Wu, D.; et al. Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme. Cancers 2020, 12, 98. https://doi.org/10.3390/cancers12010098
Lee H-H, Lin C-H, Lin H-Y, Kuei C-H, Zheng J-Q, Wang Y-H, Lu L-S, Lee F-P, Hu C-J, Wu D, et al. Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme. Cancers. 2020; 12(1):98. https://doi.org/10.3390/cancers12010098
Chicago/Turabian StyleLee, Hsun-Hua, Che-Hsuan Lin, Hui-Yu Lin, Chia-Hao Kuei, Jing-Quan Zheng, Yuan-Hung Wang, Long-Sheng Lu, Fei-Peng Lee, Chaur-Jong Hu, Dean Wu, and et al. 2020. "Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme" Cancers 12, no. 1: 98. https://doi.org/10.3390/cancers12010098





