A Comprehensive Molecular Characterization of the Pancreatic Neuroendocrine Tumor Cell Lines BON-1 and QGP-1
Abstract
:1. Introduction
2. Results
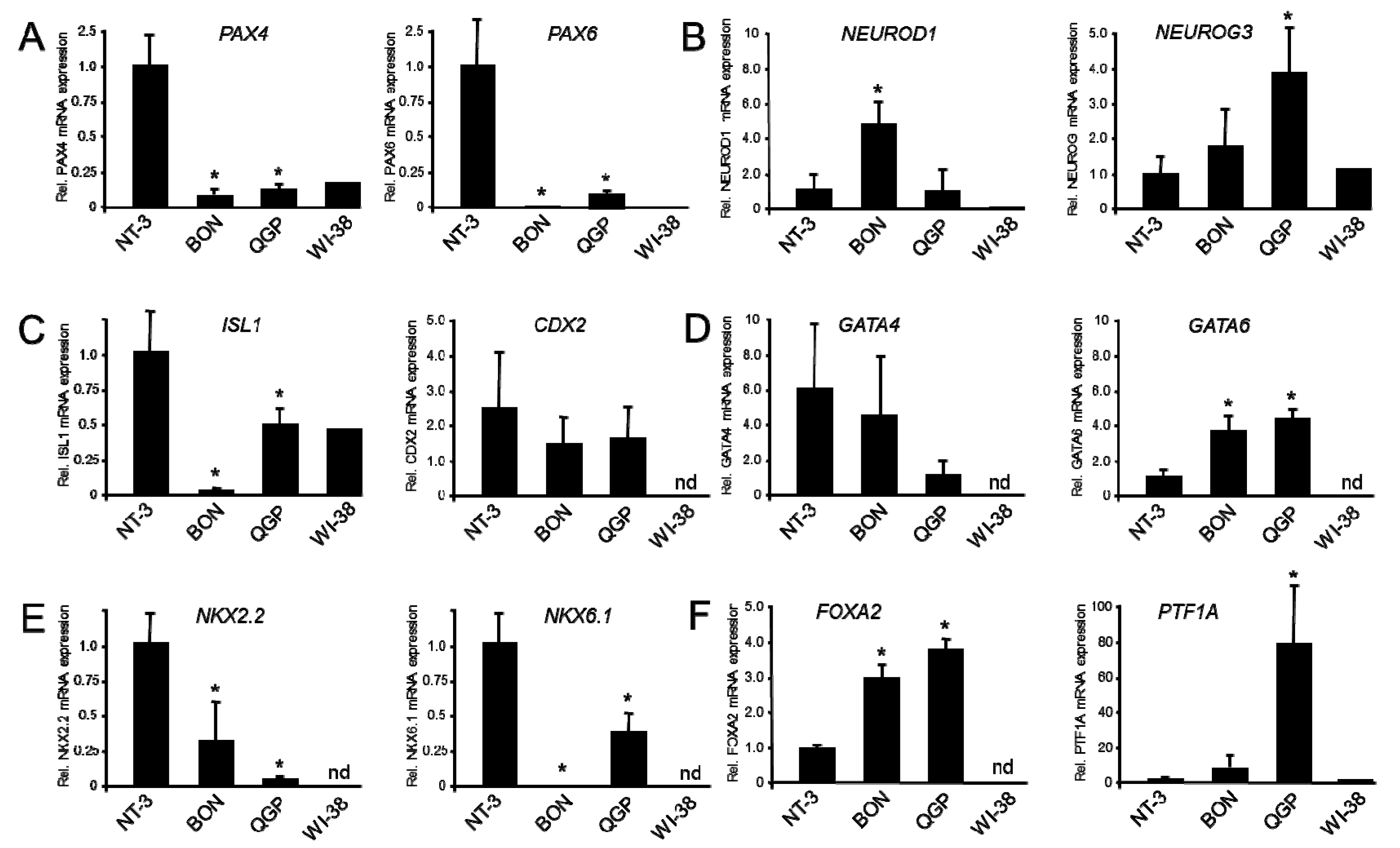
2.1. Expression of Markers of Neuroendocrine Differentiation
2.2. Expression of Markers of Epithelial/Mesenchymal Differentiation
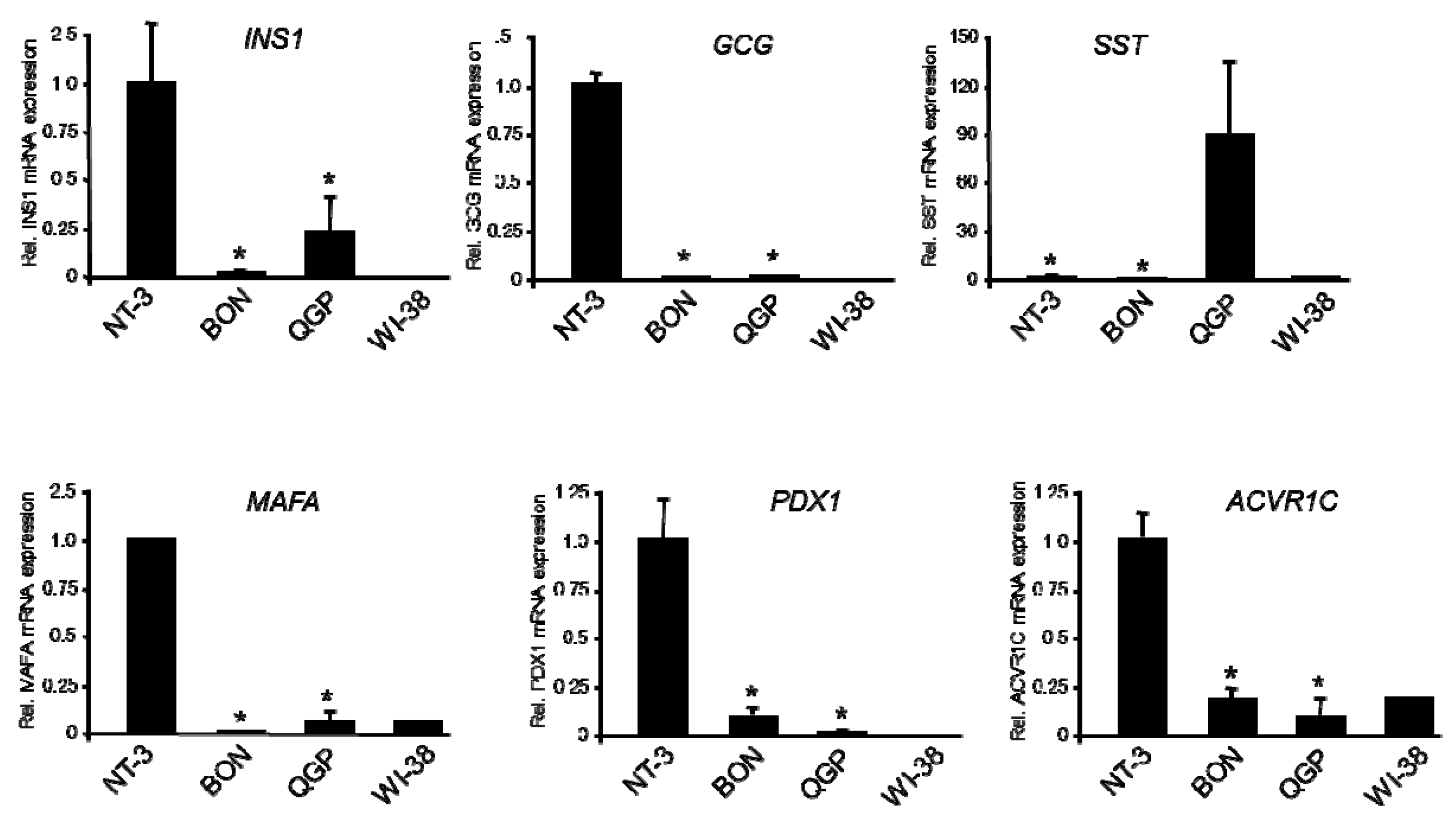
2.3. Expression of Genes of Islet Cell Hormones and Mature β-Cell Differentiation
2.4. Expression of Genes of Immature β-Cells and Lineage-Specific Pancreatic Progenitors
2.5. Identification of MiRs Expressed in BON and QGP Cells
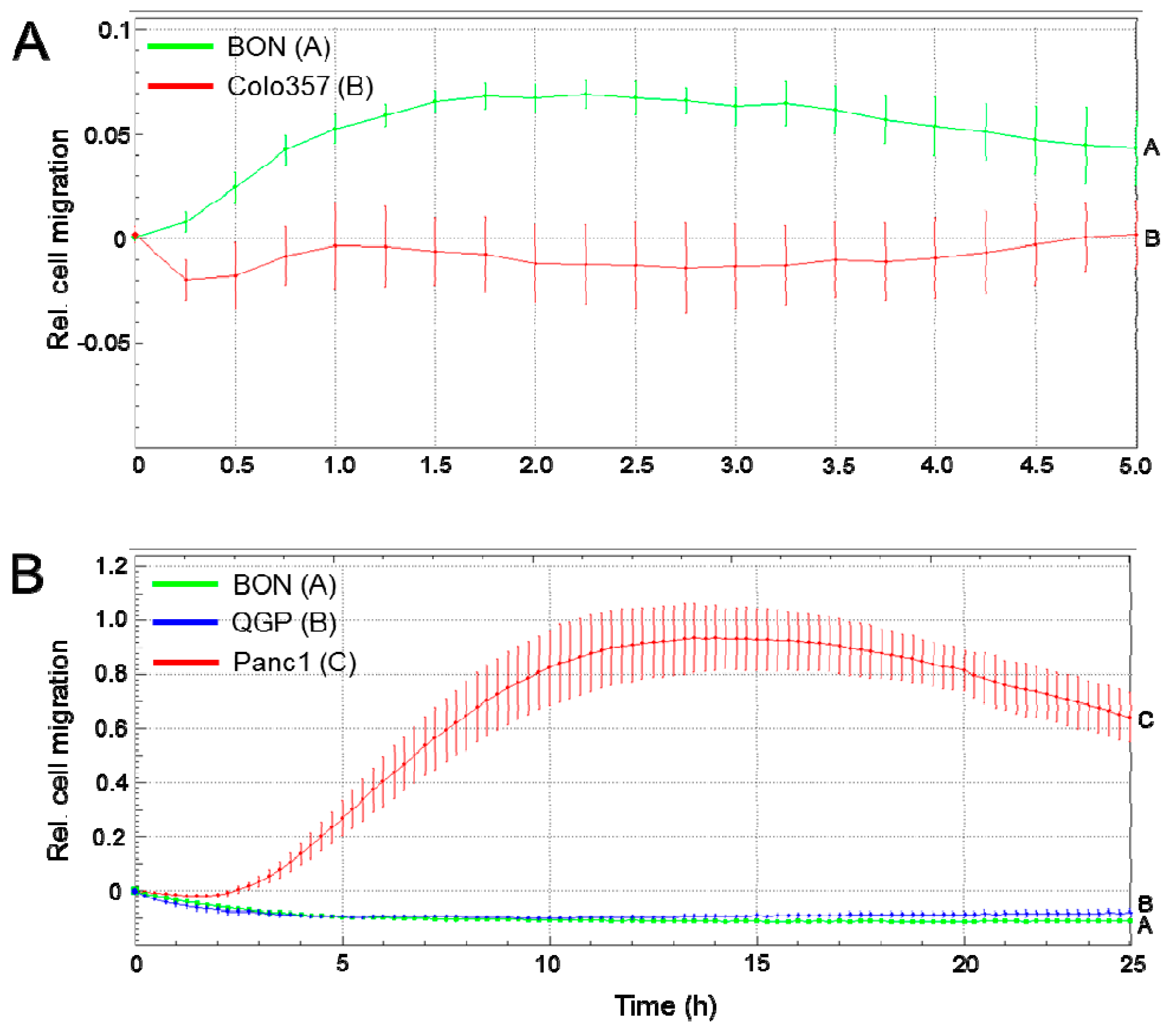
2.6. Migratory Activity of BON and QGP Cells
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Cells
5.2. RNA Isolation and Quantitative Real-Time RT-PCR (qPCR)
5.3. Immunoblot Analysis
5.4. Next Generation Sequencing of MicroRNAs
5.5. Migration Assays
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Townsend, C.M.; Ishizuka, J.; Thompson, J.C. Studies of Growth Regulation in a Neuroendocrine Cell Line. Acta Oncol. 1993, 32, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaku, M.; Nishiyama, T.; Yagawa, K.; Abe, M. Establishment of a carcinoembryonic antigen-producing cell line from human pancreatic carcinoma. GANN 1980, 71, 596–601. [Google Scholar] [PubMed]
- Iguchi, H.; Hayashi, I.; Kono, A. A somatostatin-secreting cell line established from a human pancreatic islet cell carcinoma (somatostatinoma): Release experiment and immunohistochemical study. Cancer Res. 1990, 50, 3691–3693. [Google Scholar] [PubMed]
- Gueli, N.; Toto, A.; Palmieri, G.; Carmenini, G.; Delpino, A.; Ferrini, U. In vitro growth of a cell line originated from a human insulinoma. J. Exp. Clin. Cancer Res. 1987, 6, 281–285. [Google Scholar]
- Hofving, T.; Arvidsson, Y.; Almobarak, B.; Inge, L.; Pfragner, R.; Persson, M.; Stenman, G.; Kristiansson, E.; Johanson, V.; Nilsson, O. The neuroendocrine phenotype, genomic profile and therapeutic sensitivity of GEPNET cell lines. Endocr. Relat. Cancer 2018, 25, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Boora, G.K.; Kanwar, R.; Kulkarni, A.A.; Pleticha, J.; Ames, M.; Schroth, G.; Beutler, A.S.; Banck, M.S. Exome-level comparison of primary well-differentiated neuroendocrine tumors and their cell lines. Cancer Genet. 2015, 208, 374–381. [Google Scholar] [CrossRef]
- Vandamme, T.; Peeters, M.; Dogan, F.; Pauwels, P.; Van Assche, E.; Beyens, M.; Mortier, G.; Vandeweyer, G.; de Herder, W.; Van Camp, G.; et al. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. J. Mol. Endocrinol. 2015, 54, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Exner, S.; Prasad, V.; Wiedenmann, B.; Grötzinger, C. Octreotide Does Not Inhibit Proliferation in Five Neuroendocrine Tumor Cell Lines. Front. Endocrinol. (Lausanne) 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Benten, D.; Behrang, Y.; Unrau, L.; Weissmann, V.; Wolters-Eisfeld, G.; Burdak-Rothkamm, S.; Stahl, F.R.; Anlauf, M.; Grabowski, P.; Möbs, M.; et al. Establishment of the First Well-differentiated Human Pancreatic Neuroendocrine Tumor Model. Mol. Cancer Res. 2018, 16, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Thorns, C.; Schurmann, C.; Gebauer, N.; Wallaschofski, H.; Kümpers, C.; Bernard, V.; Feller, A.C.; Keck, T.; Habermann, J.K.; Begum, N.; et al. Global microRNA profiling of pancreatic neuroendocrine neoplasias. Anticancer Res. 2014, 34, 2249–2254. [Google Scholar]
- Zimmermann, N.; Knief, J.; Kacprowski, T.; Lazar-Karsten, P.; Keck, T.; Billmann, F.; Schmid, S.; Luley, K.; Lehnert, H.; Brabant, G.; et al. MicroRNA analysis of gastroenteropancreatic neuroendocrine tumors and metastases. Oncotarget 2018, 9, 28379–29390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadanandam, A.; Wullschleger, S.; Lyssiotis, C.A.; Grötzinger, C.; Barbi, S.; Bersani, S.; Körner, J.; Wafy, I.; Mafficini, A.; Lawlor, R.T.; et al. A Cross-Species Analysis in Pancreatic Neuroendocrine Tumors Reveals Molecular Subtypes with Distinctive Clinical, Metastatic, Developmental, and Metabolic Characteristics. Cancer Discov. 2015, 5, 1296–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, H.L.; Grapin-Botton, A. The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 2017, 66, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Michael, I.P.; Saghafinia, S.; Tichet, M.; Zangger, N.; Marinoni, I.; Perren, A.; Hanahan, D. ALK7 Signaling Manifests a Homeostatic Tissue Barrier That Is Abrogated during Tumorigenesis and Metastasis. Dev. Cell 2019, 49, 409–424. [Google Scholar] [CrossRef]
- Druelle, N.; Vieira, A.; Shabro, A.; Courtney, M.; Mondin, M.; Rekima, S.; Napolitano, T.; Silvano, S.; Navarro-Sanz, S.; Hadzic, B.; et al. Ectopic expression of Pax4 in pancreatic δ cells results in β-like cell neogenesis. J. Cell Biol. 2017, 216, 4299–4311. [Google Scholar] [CrossRef] [Green Version]
- Cejas, P.; Drier, Y.; Dreijerink, K.M.A.; Brosens, L.A.A.; Deshpande, V.; Epstein, C.B.; Conemans, E.B.; Morsink, F.H.M.; Graham, M.K.; Valk, G.D.; et al. Enhancer signatures stratify and predict outcomes of non-functional pancreatic neuroendocrine tumors. Nat. Med. 2019, 25, 1260–1265. [Google Scholar] [CrossRef]
- Chakraborty, C.; George Priya Doss, C.; Bandyopadhyay, S. miRNAs in insulin resistance and diabetes-associated pancreatic cancer: The ‘minute and miracle’ molecule moving as a monitor in the ‘genomic galaxy’. Curr. Drug Targets 2013, 14, 1110–1117. [Google Scholar] [CrossRef]
- Zatelli, M.C.; Grossrubatscher, E.M.; Guadagno, E.; Sciammarella, C.; Faggiano, A.; Colao, A. Circulating tumor cells and miRNAs as prognostic markers in neuroendocrine neoplasms. Endocr. Relat. Cancer 2017, 24, R223–R237. [Google Scholar] [CrossRef]
- Mandelbaum, A.D.; Kredo-Russo, S.; Aronowitz, D.; Myers, N.; Yanowski, E.; Klochendler, A.; Swisa, A.; Dor, Y.; Hornstein, E. miR-17-92 and miR-106b-25 clusters regulate beta cell mitotic checkpoint and insulin secretion in mice. Diabetologia 2019, 62, 1653–1666. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, L.; Wan, S.; Xie, Y.; Chen, X.; Ji, X.; Zhao, Q.; Wang, C.; Zhang, K.; Hock, J.M.; et al. MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol. Cell. Endocrinol. 2016, 437, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, T.; Wang, L.; Wu, J.; Guo, S. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic β cells via the inhibition of SOCS3. Int. J. Mol. Med. 2016, 38, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Zhang, L.Q.; Ding, L.; Wang, F.; Sun, Y.J.; An, Y.; Zhao, Y.; Li, Y.H.; Teng, C.B. MicroRNA-19b downregulates insulin 1 through targeting transcription factor NeuroD1. FEBS Lett. 2011, 585, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- Setyowati Karolina, D.; Sepramaniam, S.; Tan, H.Z.; Armugam, A.; Jeyaseelan, K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013, 10, 1365–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Shen, C.A.; Zhu, B.W.; An, H.Y.; Zheng, B.; Xu, S.B.; Sun, J.C.; Sun, P.C.; Zhang, W.; Wang, J.; et al. Insight into miRNAs related with glucometabolic disorder. Biomed. Pharmacother. 2019, 111, 657–665. [Google Scholar] [CrossRef]
- Melkman-Zehavi, T.; Oren, R.; Kredo-Russo, S.; Shapira, T.; Mandelbaum, A.D.; Rivkin, N.; Nir, T.; Lennox, K.A.; Behlke, M.A.; Dor, Y.; et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 2011, 30, 835–845. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Guo, S.; Li, W.; Yu, P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015, 5, 12453. [Google Scholar] [CrossRef]
- Sun, L.L.; Jiang, B.G.; Li, W.T.; Zou, J.J.; Shi, Y.Q.; Liu, Z.M. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res. Clin. Pract. 2011, 91, 94–100. [Google Scholar] [CrossRef]
- Joglekar, M.V.; Parekh, V.S.; Mehta, S.; Bhonde, R.R.; Hardikar, A.A. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev. Biol. 2007, 311, 603–612. [Google Scholar] [CrossRef] [Green Version]
- Eliasson, L. The small RNA miR-375—A pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Mol. Cell. Endocrinol. 2017, 456, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wu, J. Feud or Friend? The Role of the miR-17-92 Cluster in Tumorigenesis. Curr. Genom. 2010, 11, 129–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med. Sci. 2018, 15, 1443–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quattrochi, B.; Gulvady, A.; Driscoll, D.R.; Sano, M.; Klimstra, D.S.; Turner, C.E.; Lewis, B.C. MicroRNAs of the mir-17~92 cluster regulate multiple aspects of pancreatic tumor development and progression. Oncotarget 2017, 8, 35902–35918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Wu, J.; Liu, W.; Zuo, Y.; Chen, S.; Zhang, S.; Zeng, M.; Huang, W. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum. Gene Ther. 2010, 21, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Z.H.; Liu, X.; Liu, G.X.; Yang, H.M.; Wu, P.F. Reduced expression of miR-3653 in glioma and its correlations with clinical progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6596–6601. [Google Scholar] [PubMed]
- Zhu, W.; Luo, X.; Fu, H.; Liu, L.; Sun, P.; Wang, Z. MiR-3653 inhibits the metastasis and epithelial–mesenchymal transition of colon cancer by targeting Zeb2. Pathol. Res. Pract. 2019, 215, 152577. [Google Scholar] [CrossRef]
- Gill, P.; Kim, E.; Chua, T.C.; Clifton-Bligh, R.J.; Nahm, C.B.; Mittal, A.; Gill, A.J.; Samra, J.S. MiRNA-3653 Is a Potential Tissue Biomarker for Increased Metastatic Risk in Pancreatic Neuroendocrine Tumours. Endocr. Pathol. 2019, 30, 128–133. [Google Scholar] [CrossRef]
- Otterbein, H.; Lehnert, H.; Ungefroren, H. Negative control of cell migration by Rac1b in highly metastatic pancreatic cancer cells is mediated by sequential induction of non-activated Smad3 and biglycan. Cancers (Basel) 2019, 11, E1959. [Google Scholar] [CrossRef] [Green Version]
- Qiao, X.W.; Qiu, L.; Chen, Y.J.; Meng, C.T.; Sun, Z.; Bai, C.M.; Zhao, D.C.; Zhang, T.P.; Zhao, Y.P.; Song, Y.L.; et al. Chromogranin A is a reliable serum diagnostic biomarker for pancreatic neuroendocrine tumors but not for insulinomas. BMC Endocr. Disord. 2014, 14, 64. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Saleem, S.; Yee, S.-P.; Hardikar, A.A.; Wang, R. c-Kit and stem cell factor regulate PANC-1 cell differentiation into insulin- and glucagon-producing cells. Lab. Investig. 2010, 90, 1373–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinn, R.; Otterbein, H.; Lehnert, H.; Ungefroren, H. RAC1B: A guardian of the epithelial phenotype and protector against epithelial–mesenchymal transition. Cells 2019, 8, E1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellner, U.; Brabletz, T.; Keck, T. ZEB1 in Pancreatic Cancer. Cancers (Basel) 2010, 2, 1617–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaviani, M.; Azarpira, N.; Karimi, M.H.; Al-Abdullah, I. The role of microRNAs in islet β-cell development. Cell Biol. Int. 2016, 40, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- van de Bunt, M.; Gaulton, K.J.; Parts, L.; Moran, I.; Johnson, P.R.; Lindgren, C.M.; Ferrer, J.; Gloyn, A.L.; McCarthy, M.I. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS ONE 2013, 8, e55272. [Google Scholar] [CrossRef] [PubMed]
- Esguerra, J.L.S.; Nagao, M.; Ofori, J.K.; Wendt, A.; Eliasson, L. MicroRNAs in islet hormone secretion. Diabetes Obes. Metab. 2018, 20 (Suppl. 2), 11–19. [Google Scholar] [CrossRef] [Green Version]
- Roldo, C.; Missiaglia, E.; Hagan, J.P.; Falconi, M.; Capelli, P.; Bersani, S.; Calin, G.A.; Volinia, S.; Liu, C.G.; Scarpa, A.; et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006, 24, 4677–4684. [Google Scholar] [CrossRef]
- Ryu, J.K.; Matthaei, H.; Dal Molin, M.; Hong, S.M.; Canto, M.I.; Schulick, R.D.; Wolfgang, C.; Goggins, M.G.; Hruban, R.H.; Cope, L.; et al. Elevated microRNA miR-21 levels in pancreatic cyst fluid are predictive of mucinous precursor lesions of ductal adenocarcinoma. Pancreatology 2011, 11, 343–350. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Li, Y.; Zhao, Y.; Zhai, W.; Yang, L.; Kong, D.; Wu, C.; Chen, Z.; Teng, C.B. miR-18a counteracts AKT and ERK activation to inhibit the proliferation of pancreatic progenitor cells. Sci. Rep. 2017, 7, 45002. [Google Scholar] [CrossRef] [Green Version]
- Passacantilli, I.; Capurso, G.; Archibugi, L.; Calabretta, S.; Caldarola, S.; Loreni, F.; Delle Fave, G.; Sette, C. Combined therapy with RAD001 e BEZ235 overcomes resistance of PET immortalized cell lines to mTOR inhibition. Oncotarget 2014, 5, 5381–5391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.C.; Pavel, M.; Lombard-Bohas, C.; Van Cutsem, E.; Voi, M.; Brandt, U.; He, W.; Chen, D.; Capdevila, J.; de Vries, E.G.E.; et al. Everolimus for the Treatment of Advanced Pancreatic Neuroendocrine Tumors: Overall Survival and Circulating Biomarkers from the Randomized, Phase III RADIANT-3 Study. J. Clin. Oncol. 2016, 34, 3906–3913. [Google Scholar] [CrossRef] [PubMed]
- Di Florio, A.; Adesso, L.; Pedrotti, S.; Capurso, G.; Pilozzi, E.; Corbo, V.; Scarpa, A.; Geremia, R.; Delle Fave, G.; Sette, C. Src kinase activity coordinates cell adhesion and spreading with activation of mammalian target of rapamycin in pancreatic endocrine tumour cells. Endocr. Relat. Cancer 2011, 18, 541–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Florio, A.; Capurso, G.; Milione, M.; Panzuto, F.; Geremia, R.; Delle Fave, G.; Sette, C. Src family kinase activityregulates adhesion, spreading and migration of pancreatic endocrine tumour cells. Endocr. Relat. Cancer 2007, 14, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Schmalhofer, O.; Brabletz, S.; Brabletz, T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009, 28, 151–166. [Google Scholar] [CrossRef]
- Detjen, K.M.; Rieke, S.; Deters, A.; Schulz, P.; Rexin, A.; Vollmer, S.; Hauff, P.; Wiedenmann, B.; Pavel, M.; Scholz, A. Angiopoietin-2 promotes disease progression of neuroendocrine tumors. Clin. Cancer Res. 2010, 16, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Scholz, A.; Wagner, K.; Welzel, M.; Remlinger, F.; Wiedenmann, B.; Siemeister, G.; Rosewicz, S.; Detjen, K.M. The oral multitarget tumour growth inhibitor, ZK 304709, inhibits growth of pancreatic neuroendocrine tumours in an orthotopic mouse model. Gut 2009, 58, 261–270. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y.; Li, J.; Ma, D. MicroRNA-664 Targets Insulin Receptor Substrate 1 to Suppress Cell Proliferation and Invasion in Breast Cancer. Oncol. Res. 2019, 27, 459–467. [Google Scholar] [CrossRef]
- Guo, S.; Xu, X.; Tang, Y.; Zhang, C.; Li, J.; Ouyang, Y.; Ju, J.; Bie, P.; Wang, H. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett. 2014, 344, 40–46. [Google Scholar] [CrossRef]
- Michael, I.P.; Saghafinia, S.; Hanahan, D. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc. Natl. Acad. Sci. USA 2019, 116, 24184–24195. [Google Scholar] [CrossRef] [Green Version]
- Evers, B.M.; Townsend, C.M., Jr.; Upp, J.R.; Allen, E.; Hurlbut, S.C.; Kim, S.W.; Rajaraman, S.; Singh, P.; Reubi, J.C.; Thompson, J.C. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology 1991, 101, 303–311. [Google Scholar] [CrossRef]
- Lopez, J.R.; Claessen, S.M.; Macville, M.V.; Albrechts, J.C.; Skogseid, B.; Speel, E.J. Spectral karyotypic and comparative genomic analysis of the endocrine pancreatic tumor cell line BON-1. Neuroendocrinology 2010, 91, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Cataldo Bascuñan, L.R.; Lyons, C.; Bennet, H.; Artner, I.; Fex, M. Serotonergic regulation of insulin secretion. Acta Physiol. 2019, 225, e13101. [Google Scholar] [CrossRef]
- Gradiz, R.; Silva, H.C.; Carvalho, L.; Botelho, M.F.; Mota-Pinto, A. MIA PaCa-2 and PANC-1—Pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci. Rep. 2016, 6, 21648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Tedesco, L.; Lucia, K.; Schlitter, A.M.; Garcia, J.M.; Esposito, I.; Auernhammer, C.J.; Theodoropoulou, M.; Arzt, E.; Renner, U.; et al. RSUME is implicated in tumorigenesis and metastasis of pancreatic neuroendocrine tumors. Oncotarget 2016, 7, 57878–57893. [Google Scholar] [CrossRef] [PubMed]
- Hamil, L.; Benghuzzi, H.; Tucci, M. Evaluation of insulin secretion by pancreatic cells in response to increasing amounts of glucose. Biomed. Sci. Instrum. 2008, 44, 441–446. [Google Scholar]
- Donadel, G.; Pastore, D.; Della-Morte, D.; Capuani, B.; Lombardo, M.F.; Pacifici, F.; Bugliani, M.; Grieco, F.A.; Marchetti, P.; Lauro, D. FGF-2b and h-PL Transform Duct and Non-Endocrine Human Pancreatic Cells into Endocrine Insulin Secreting Cells by Modulating Differentiating Genes. Int. J. Mol. Sci. 2017, 18, E2234. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wang, H.; Saunee, N.A.; Breslin, M.B.; Lan, M.S. Insulinoma-associated antigen-1 zinc-finger transcription factor promotes pancreatic duct cell trans-differentiation. Endocrinology 2010, 151, 2030–2039. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luley, K.B.; Biedermann, S.B.; Künstner, A.; Busch, H.; Franzenburg, S.; Schrader, J.; Grabowski, P.; Wellner, U.F.; Keck, T.; Brabant, G.; et al. A Comprehensive Molecular Characterization of the Pancreatic Neuroendocrine Tumor Cell Lines BON-1 and QGP-1. Cancers 2020, 12, 691. https://doi.org/10.3390/cancers12030691
Luley KB, Biedermann SB, Künstner A, Busch H, Franzenburg S, Schrader J, Grabowski P, Wellner UF, Keck T, Brabant G, et al. A Comprehensive Molecular Characterization of the Pancreatic Neuroendocrine Tumor Cell Lines BON-1 and QGP-1. Cancers. 2020; 12(3):691. https://doi.org/10.3390/cancers12030691
Chicago/Turabian StyleLuley, Kim B., Shauni B. Biedermann, Axel Künstner, Hauke Busch, Sören Franzenburg, Jörg Schrader, Patricia Grabowski, Ulrich F. Wellner, Tobias Keck, Georg Brabant, and et al. 2020. "A Comprehensive Molecular Characterization of the Pancreatic Neuroendocrine Tumor Cell Lines BON-1 and QGP-1" Cancers 12, no. 3: 691. https://doi.org/10.3390/cancers12030691




