The Plasma Membrane Ca2+ Pump PMCA4b Regulates Melanoma Cell Migration through Remodeling of the Actin Cytoskeleton
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Chemicals and Reagents
2.3. DNA Constructs
2.4. Generation of Stable Cell Lines
2.5. Transient Transfection
2.6. siRNA Transfection
2.7. Cell Morphology Analysis
2.8. Nearest Neighbour Distance Analysis
2.9. Non-Directional Cell Motility Assay
2.10. Directional Cell Migration Assay
2.11. Immunofluorescence Microscopy
2.12. Live-Cell Imaging
2.13. Fluorescence Recovery after Photobleaching (FRAP)
2.14. Ca2+ Signal Measurements
2.15. Western Blot Analysis
2.16. Statistical Analysis
3. Results
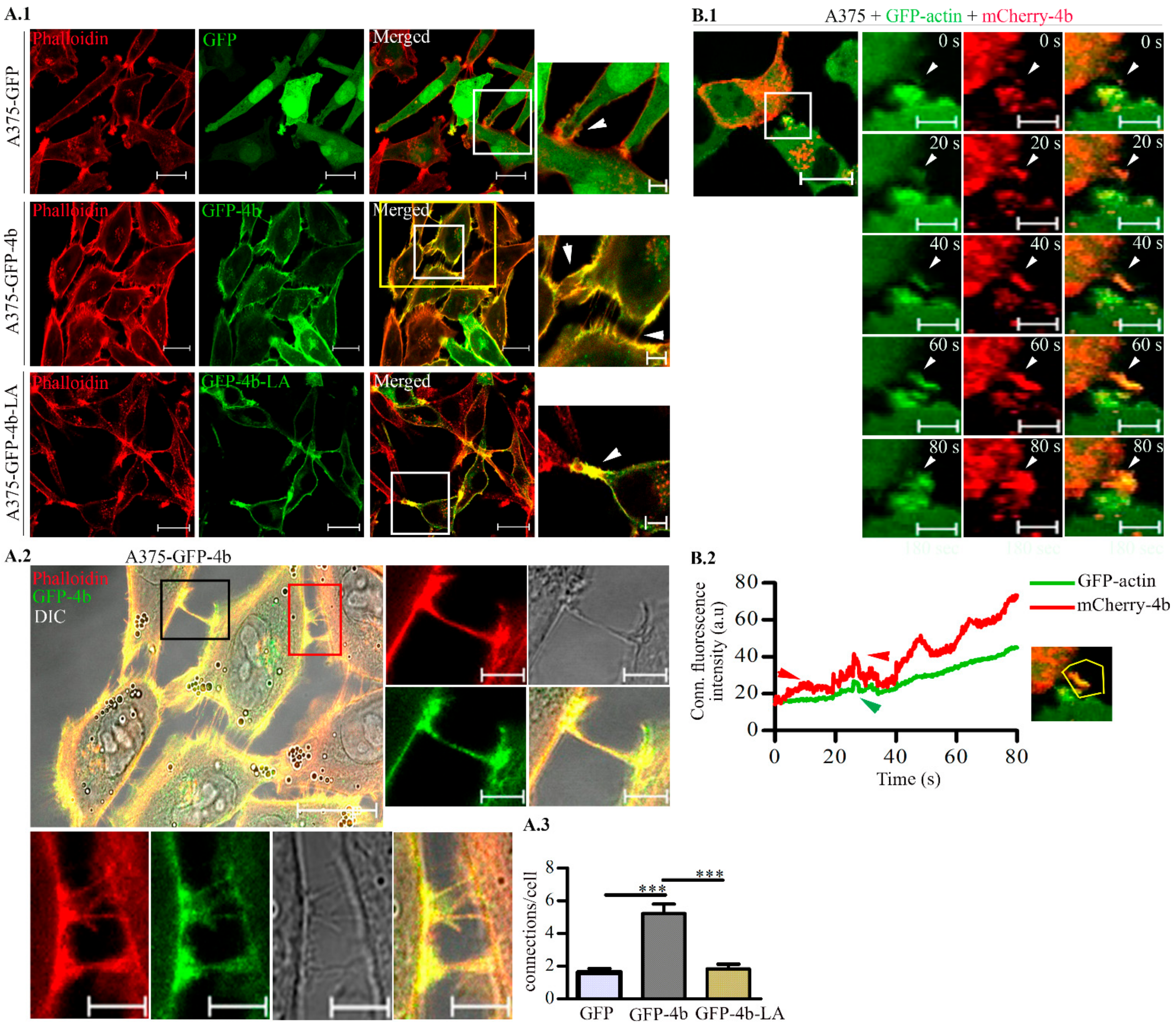
3.1. Proper Trafficking of PMCA4b Is Required to Change A375 Melanoma Cell Morphology, and Migration.
3.2. PMCA4b Induces Actin Cytoskeleton Remodeling in A375 Melanoma Cells
3.3. A Functional PMCA4b Pump Is Needed for Actin Cytoskeleton Remodeling
3.4. Silencing PMCA4b Expression Decreases the Number of Cells with Stress Fibers and Changes Cell Culture Morphology of MCF-7 Breast Cancer Cells
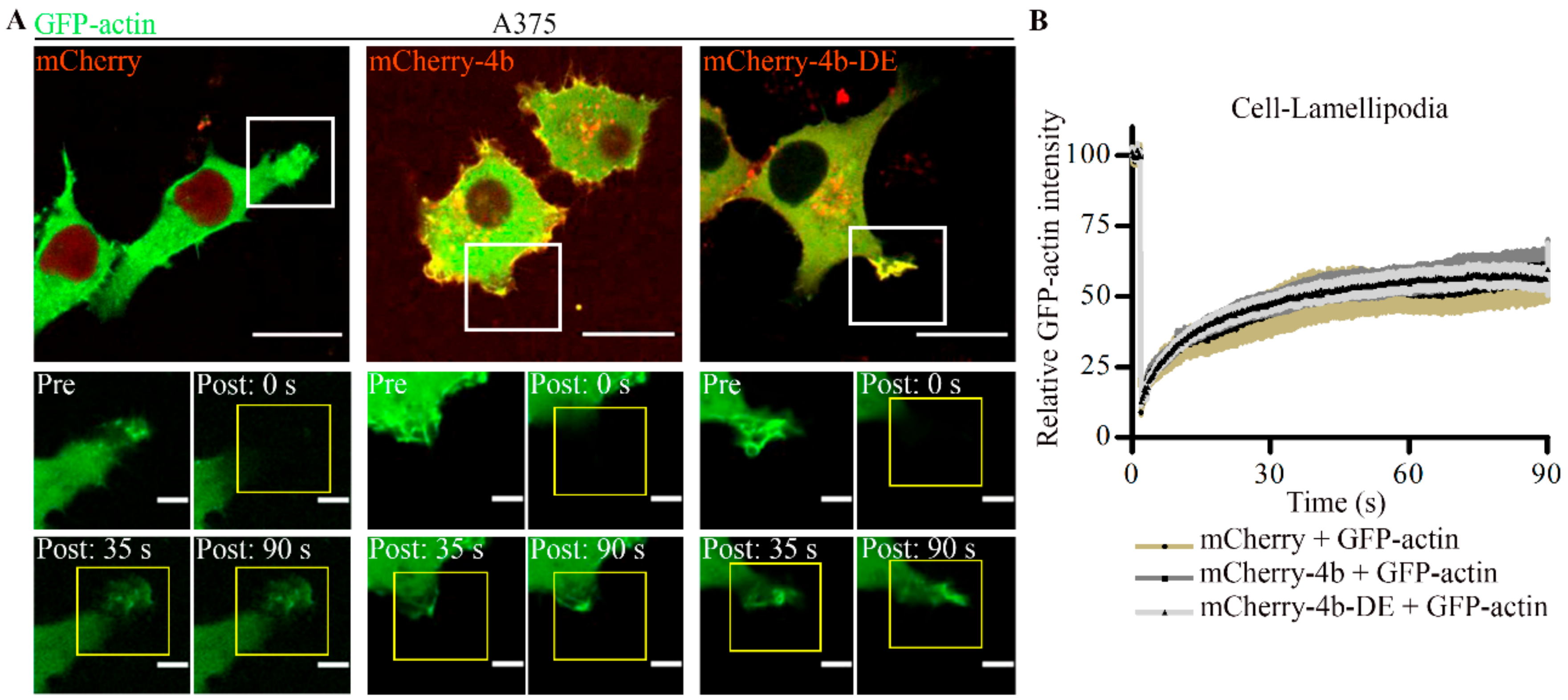
3.5. PMCA4b Does Not Affect F-Actin Recovery after Photobleaching
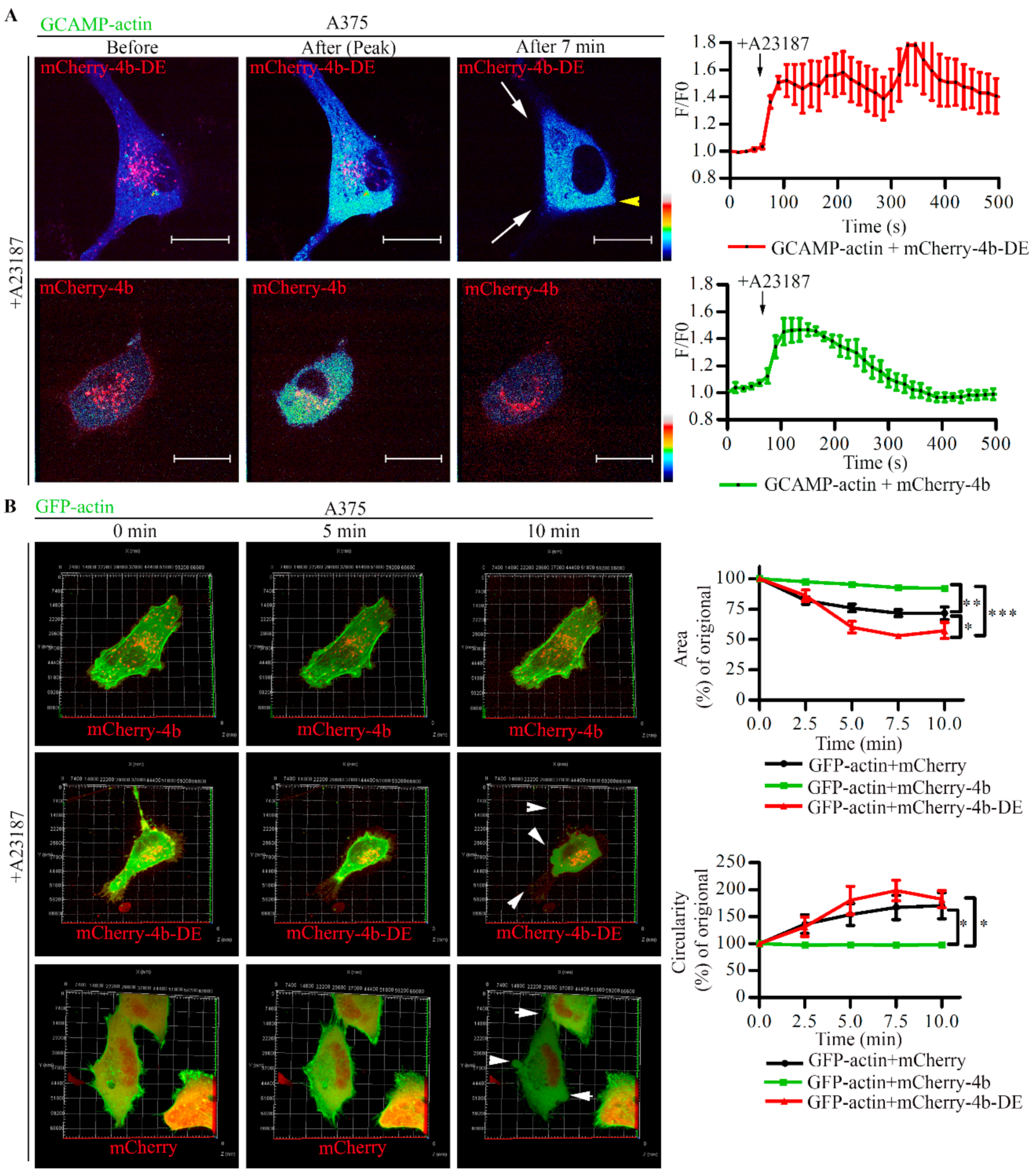
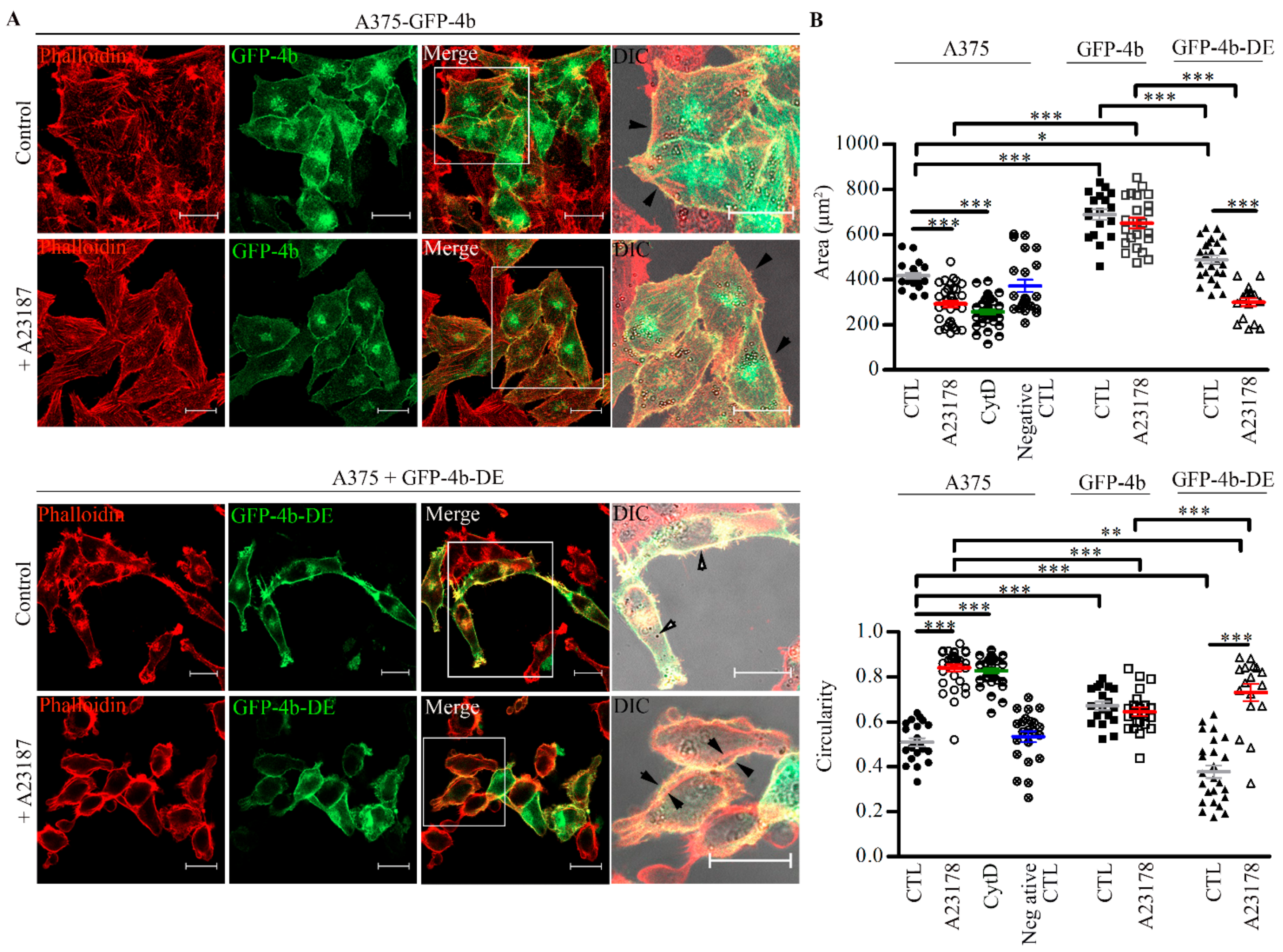
3.6. PMCA4b Inhibits Ca2+ Induced F-Actin Depolymerization
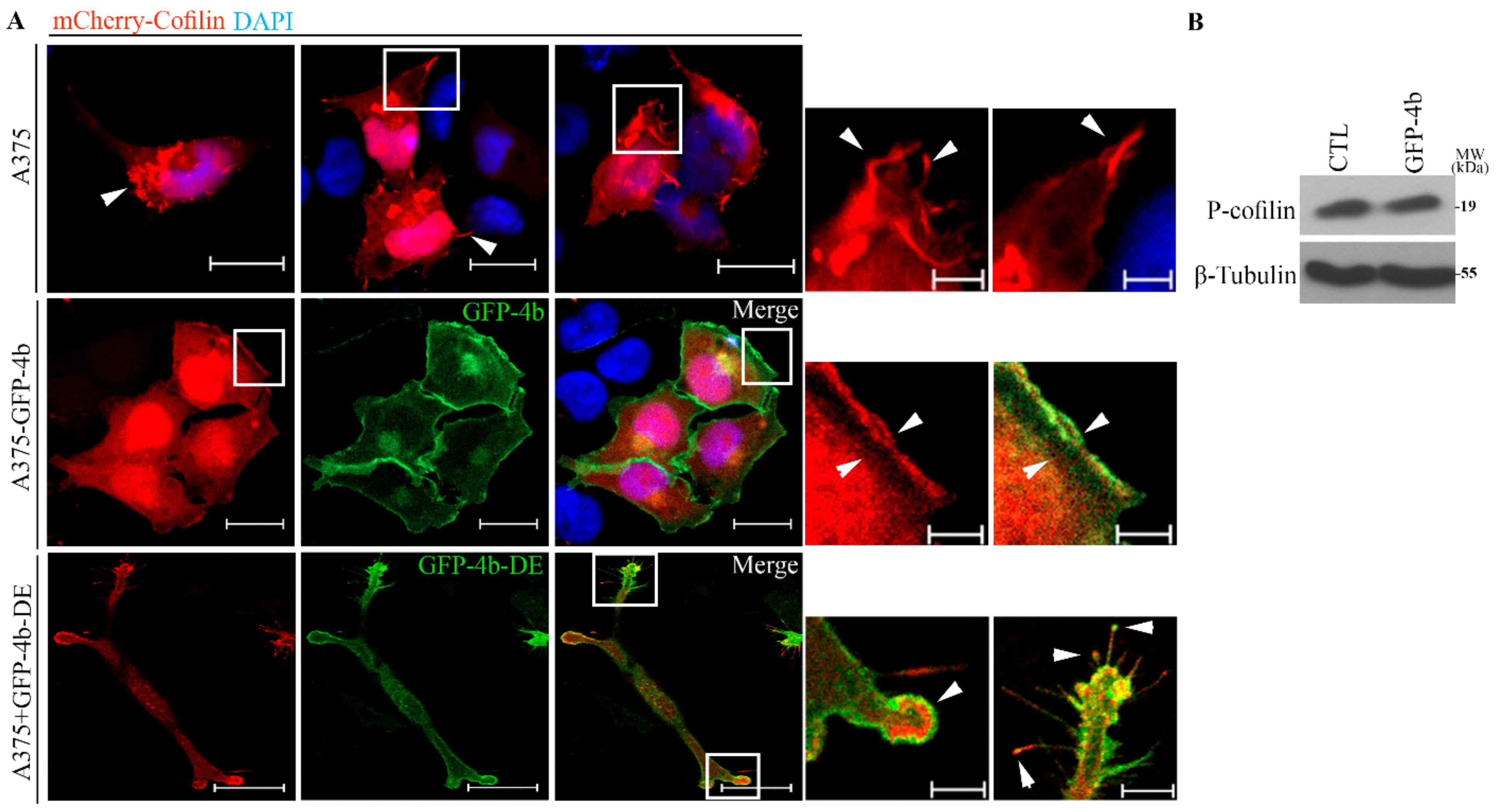
3.7. PMCA4b Induces F-Actin Rearrangement through Cofilin Relocation.
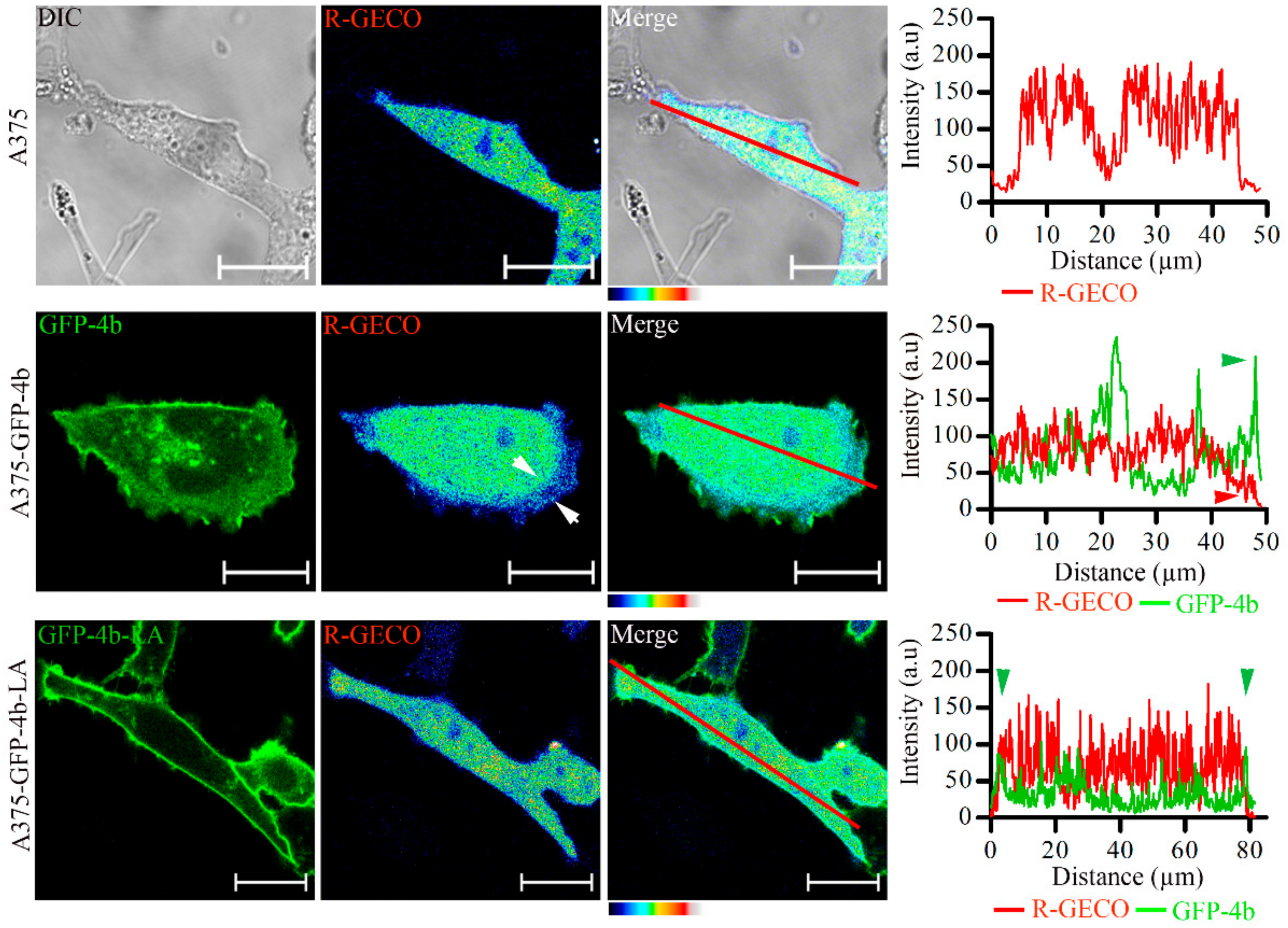
3.8. Proper PMCA4b Trafficking Is Essential in Managing Front-to-Rear Increasing Ca2+ Concentration Gradient in A375 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adler, N.R.; Haydon, A.; McLean, C.A.; Kelly, J.W.; Mar, V.J. Metastatic pathways in patients with cutaneous melanoma. Pigment. Cell Melanoma Res. 2017, 30, 13–27. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.Z.; Garcia, M.V.; Li, T.Y.; Khor, L.Y.; Gajapathy, R.S.; Spittle, C.; Weed, S.; Lessin, S.R.; Wu, H. Cytoskeleton alterations in melanoma: Aberrant expression of cortactin, an actin-binding adapter protein, correlates with melanocytic tumor progression. Mod. Pathol. 2010, 23, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Pawlak, G.; Helfman, D.M. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 2001, 11, 41–47. [Google Scholar] [CrossRef]
- Maus, M.; Medgyesi, D.; Kiss, E.; Schneider, A.E.; Enyedi, A.; Szilágyi, N.; Matkó, J.; Sármay, G. B cell receptor-induced Ca2+ mobilization mediates F-actin rearrangements and is indispensable for adhesion and spreading of B lymphocytes. J. Leukoc. Biol. 2013, 93, 537–547. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Ishihara, A.; Oxford, G.; Johnson, B.; Jacobson, K. Regulation of cell movement is mediated by stretch-activated calcium channels. Nature 1999, 400, 382–386. [Google Scholar] [CrossRef]
- Brundage, R.A.; Fogarty, K.E.; Tuft, R.A.; Fay, F.S. Calcium gradients underlying polarization and chemotaxis of eosinophils. Science 1991, 254, 703–706. [Google Scholar] [CrossRef]
- Martin-Romero, F.J.; Lopez-Guerrero, A.M.; Pascual-Caro, C.; Pozo-Guisado, E. The Interplay between Cytoskeleton and Calcium Dynamics. In Cytoskeleton—Structure, Dynamics, Function and Disease; Jimenez-Lopez, J.C., Ed.; IntechOpen: Rijeka, Croatia, 2017; Chapter 4; pp. 73–88. [Google Scholar]
- Tsai, F.C.; Kuo, G.H.; Chang, S.W.; Tsai, P.J. Ca2+ signaling in cytoskeletal reorganization, cell migration, and cancer metastasis. Biomed. Res. Int. 2015, 2015, 409245. [Google Scholar] [CrossRef] [Green Version]
- Marchi, S.; Pinton, P. Alterations of calcium homeostasis in cancer cells. Curr. Opin. Pharmacol. 2016, 29, 1–6. [Google Scholar] [CrossRef]
- Déliot, N.; Constantin, B. Plasma membrane calcium channels in cancer: Alterations and consequences for cell proliferation and migration. Biochim. Biophys. Acta 2015, 1848, 2512–2522. [Google Scholar] [CrossRef] [Green Version]
- Maiques, O.; Barceló, C.; Panosa, A.; Pijuan, J.; Orgaz, J.L.; Rodriguez-Hernandez, I.; Matas-Nadal, C.; Tell, G.; Vilella, R.; Fabra, A.; et al. T-type calcium channels drive migration/invasion in BRAFV600E melanoma cells through Snail1. Pigment. Cell Melanoma Res. 2018, 31, 484–495. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Cao, R.; Qian, K.; Peng, T.; Yuan, L.; Chen, L.; Cheng, S.; Xiong, Y.; Ju, L.; Wang, X.; et al. TRPM8 inhibition regulates the proliferation, migration and ROS metabolism of bladder cancer cells. OncoTargets Ther. 2020, 13, 8825–8835. [Google Scholar] [CrossRef]
- Monet, M.; Lehen’kyi, V.; Gackiere, F.; Firlej, V.; Vandenberghe, M.; Roudbaraki, M.; Gkika, D.; Pourtier, A.; Bidaux, G.; Slomianny, C.; et al. Role of cationic channel TRPV2 in promoting prostate cancer migration and progression to androgen resistance. Cancer Res. 2010, 70, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, J. Mechanical tumor microenvironment and transduction: Cytoskeleton mediates cancer cell invasion and metastasis. Int. J. Biol. Sci. 2020, 16, 2014–2028. [Google Scholar] [CrossRef]
- Shimizu, T.; Owsianik, G.; Freichel, M.; Flockerzi, V.; Nilius, B.; Vennekens, R. TRPM4 regulates migration of mast cells in mice. Cell Calcium 2009, 45, 226–232. [Google Scholar] [CrossRef]
- Lee, W.H.; Choong, L.Y.; Mon, N.N.; Lu, S.; Lin, Q.; Pang, B.; Yan, B.; Krishna, V.S.; Singh, H.; Tan, T.Z.; et al. TRPV4 regulates breast cancer cell extravasation, stiffness and actin cortex. Sci. Rep. 2016, 6, 27903. [Google Scholar] [CrossRef] [Green Version]
- Pinotsis, N.; Zielinska, K.; Babuta, M.; Arolas, J.L.; Kostan, J.; Khan, M.B.; Schreiner, C.; Salmazo, A.; Ciccarelli, L.; Puchinger, M.; et al. Calcium modulates the domain flexibility and function of an α-actinin similar to the ancestral α-actinin. Proc. Natl. Acad. Sci. USA 2020, 117, 22101–22112. [Google Scholar] [CrossRef]
- Oser, M.; Condeelis, J. The cofilin activity cycle in lamellipodia and invadopodia. J. Cell Biochem. 2009, 108, 1252–1262. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Cordero, J.J.; Magalhaes, M.A.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shibasaki, F.; Mizuno, K. Calcium signal-induced cofilin dephosphorylation is mediated by Slingshot via calcineurin. J. Biol. Chem. 2005, 280, 12683–12689. [Google Scholar] [CrossRef] [Green Version]
- Padányi, R.; Pászty, K.; Hegedűs, L.; Varga, K.; Papp, B.; Penniston, J.T.; Enyedi, Á. Multifaceted plasma membrane Ca(2+) pumps: From structure to intracellular Ca(2+) handling and cancer. Biochim. Biophys. Acta 2016, 1863, 1351–1363. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Van Houten, J.N.; Dann, P.; Kim, W.; Sullivan, C.; Yu, H.; Liotta, L.; Espina, V.; Stern, D.F.; Friedman, P.A.; et al. PMCA2 regulates HER2 protein kinase localization and signaling and promotes HER2-mediated breast cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E282–E290. [Google Scholar] [CrossRef] [Green Version]
- Rüschoff, J.H.; Brandenburger, T.; Strehler, E.E.; Filoteo, A.G.; Heinmöller, E.; Aumüller, G.; Wilhelm, B. Plasma membrane calcium ATPase expression in human colon multistep carcinogenesis. Cancer Investig. 2012, 30, 251–257. [Google Scholar] [CrossRef]
- Dhanasekaran, S.M.; Barrette, T.R.; Ghosh, D.; Shah, R.; Varambally, S.; Kurachi, K.; Pienta, K.J.; Rubin, M.A.; Chinnaiyan, A.M. Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412, 822–826. [Google Scholar] [CrossRef]
- Hegedũs, L.; Garay, T.; Molnár, E.; Varga, K.; Bilecz, Á.; Török, S.; Padányi, R.; Pászty, K.; Wolf, M.; Grusch, M.; et al. The plasma membrane Ca2+ pump PMCA4b inhibits the migratory and metastatic activity of BRAF mutant melanoma cells. Int. J. Cancer 2017, 140, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Naffa, R.; Vogel, L.; Hegedűs, L.; Pászty, K.; Tóth, S.; Kelemen, K.; Singh, N.; Reményi, A.; Kállay, E.; Cserepes, M.; et al. P38 MAPK promotes migration and metastatic activity of BRAF mutant melanoma cells by inducing degradation of PMCA4b. Cells 2020, 9, 1209. [Google Scholar] [CrossRef]
- Hegedüs, L.; Padányi, R.; Molnár, J.; Pászty, K.; Varga, K.; Kenessey, I.; Sárközy, E.; Wolf, M.; Grusch, M.; Hegyi, Z.; et al. Histone deacetylase inhibitor treatment increases the expression of the plasma membrane Ca2+ pump PMCA4b and inhibits the migration of melanoma cells independent of ERK. Front. Oncol. 2017, 7, 95. [Google Scholar] [CrossRef] [Green Version]
- Varga, K.; Pászty, K.; Padányi, R.; Hegedűs, L.; Brouland, J.-P.; Papp, B.; Enyedi, A. Histone deacetylase inhibitor- and PMA-induced upregulation of PMCA4b enhances Ca2+ clearance from MCF-7 breast cancer cells. Cell Calcium. 2014, 55, 78–92. [Google Scholar] [CrossRef]
- Ribiczey, P.; Tordai, A.; Andrikovics, H.; Filoteo, A.G.; Penniston, J.T.; Enouf, J.; Enyedi, A.; Papp, B.; Kovács, T. Isoform-specific up-regulation of plasma membrane Ca2+ ATPase expression during colon and gastric cancer cell differentiation. Cell Calcium. 2007, 42, 590–605. [Google Scholar] [CrossRef] [Green Version]
- Antalffy, G.; Pászty, K.; Varga, K.; Hegedűs, L.; Enyedi, Á.; Padányi, R. A C-terminal di-leucine motif controls plasma membrane expression of PMCA4b. Biochim. Biophys. Acta 2013, 1833, 2561–2572. [Google Scholar] [CrossRef] [Green Version]
- Penniston, J.T.; Padányi, R.; Pászty, K.; Varga, K.; Hegedus, L.; Enyedi, A. Apart from its known function, the plasma membrane Ca2+ATPase can regulate Ca2+ signaling by controlling phosphatidylinositol 4,5-bisphosphate levels. J. Cell Sci. 2014, 127, 784. [Google Scholar] [CrossRef] [Green Version]
- Mao, T.; O’Connor, D.H.; Scheuss, V.; Nakai, J.; Svoboda, K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE 2008, 3, e1796. [Google Scholar] [CrossRef]
- Taylor, M.J.; Perrais, D.; Merrifield, C.J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011, 9, e1000604. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [Green Version]
- Traynor, D.; Kay, R.R. Possible roles of the endocytic cycle in cell motility. J. Cell Sci. 2007, 120, 2318–2327. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.J.; Allen, J.L.; Caswell, P.T. Vesicle trafficking pathways that direct cell migration in 3D matrices and in vivo. Traffic 2018, 19, 899–909. [Google Scholar] [CrossRef] [Green Version]
- Sigismund, S.; Confalonieri, S.; Ciliberto, A.; Polo, S.; Scita, G.; Di Fiore, P.P. Endocytosis and signaling: Cell logistics shape the eukaryotic cell plan. Physiol. Rev. 2012, 92, 273–366. [Google Scholar] [CrossRef] [Green Version]
- Svitkina, T. The Actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 2018, 10, a018267. [Google Scholar] [CrossRef] [Green Version]
- Franco, S.J.; Rodgers, M.A.; Perrin, B.J.; Han, J.; Bennin, D.A.; Critchley, D.R.; Huttenlocher, A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004, 6, 977–983. [Google Scholar] [CrossRef]
- Giannone, G.; Rondé, P.; Gaire, M.; Beaudouin, J.; Haiech, J.; Ellenberg, J.; Takeda, K. Calcium rises locally trigger focal adhesion disassembly and enhance residency of focal adhesion kinase at focal adhesions. J. Biol. Chem. 2004, 279, 28715–28723. [Google Scholar] [CrossRef] [Green Version]
- Pászty, K.; Caride, A.J.; Bajzer, Ž.; Offord, C.P.; Padányi, R.; Hegedűs, L.; Varga, K.; Strehler, E.E.; Enyedi, A. Plasma membrane Ca2+-ATPases can shape the pattern of Ca2+ transients induced by store-operated Ca2+ entry. Sci. Signal. 2015, 8, ra19. [Google Scholar] [CrossRef] [Green Version]
- Koskinen, M.; Hotulainen, P. Measuring F-actin properties in dendritic spines. Front. Neuroanat. 2014, 8, 74. [Google Scholar] [CrossRef] [Green Version]
- Hartzell, C.A.; Jankowska, K.I.; Burkhardt, J.K.; Lewis, R.S. Calcium influx through CRAC channels controls actin organization and dynamics at the immune synapse. eLife 2016, 5, e14850. [Google Scholar] [CrossRef]
- Yoneda, M.; Nishizaki, T.; Tasaka, K.; Kurachi, H.; Miyake, A.; Murata, Y. Changes in actin network during calcium-induced exocytosis in permeabilized GH3 cells: Calcium directly regulates F-actin disassembly. J. Endocrinol. 2000, 166, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Wales, P.; Schuberth, C.E.; Aufschnaiter, R.; Fels, J.; García-Aguilar, I.; Janning, A.; Dlugos, C.P.; Schäfer-Herte, M.; Klingner, C.; Wälte, M.; et al. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 2016, 5, e19850. [Google Scholar] [CrossRef]
- Moon, A.; Drubin, D.G. The ADF/cofilin proteins: Stimulus-responsive modulators of actin dynamics. Mol. Biol. Cell 1995, 6, 1423–1431. [Google Scholar] [CrossRef] [Green Version]
- Tsai, F.C.; Seki, A.; Yang, H.W.; Hayer, A.; Carrasco, S.; Malmersjö, S.; Meyer, T. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 2014, 16, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Hsu, M.-Y.; Meier, F.; Herlyn, M. Melanoma development and progression: A conspiracy between tumor and host. Differentiation 2002, 70, 522–536. [Google Scholar] [CrossRef]
- Elsholz, F.; Harteneck, C.; Muller, W.; Friedland, K. Calcium—A central regulator of keratinocyte differentiation in health and disease. Eur. J. Dermatol. 2014, 24, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Kodet, O.; Lacina, L.; Krejčí, E.; Dvořánková, B.; Grim, M.; Štork, J.; Kodetová, D.; Vlček, Č.; Šáchová, J.; Kolář, M.; et al. Melanoma cells influence the differentiation pattern of human epidermal keratinocytes. Mol. Cancer 2015, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Škalamera, D.; Stevenson, A.J.; Ehmann, A.; Ainger, S.A.; Lanagan, C.; Sturm, R.A.; Gabrielli, B. Melanoma mutations modify melanocyte dynamics in co-culture with keratinocytes or fibroblasts. J. Cell Sci. 2019, 132, jcs234716. [Google Scholar] [CrossRef]
- Parri, M.; Taddei, M.L.; Bianchini, F.; Calorini, L.; Chiarugi, P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 2009, 69, 2072–2081. [Google Scholar] [CrossRef] [Green Version]
- Taddei, M.L.; Giannoni, E.; Morandi, A.; Ippolito, L.; Ramazzotti, M.; Callari, M.; Gandellini, P.; Chiarugi, P. Mesenchymal to amoeboid transition is associated with stem-like features of melanoma cells. Cell Commun. Signal. 2014, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P.; Wolf, K. Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.; Harper, A.G.S. Human platelets use a cytosolic Ca2+ nanodomain to activate Ca2+-dependent shape change independently of platelet aggregation. Cell Calcium. 2020, 90, 102248. [Google Scholar] [CrossRef]
- Mermelstein, C.S.; Rebello, M.I.; Amaral, L.M.; Costa, M.L. Changes in cell shape, cytoskeletal proteins and adhesion sites of cultured cells after extracellular Ca2+ chelation. Braz. J. Med. Biol. Res. 2003, 36, 1111–1116. [Google Scholar] [CrossRef] [Green Version]
- Moore, T.M.; Brough, G.H.; Babal, P.; Kelly, J.J.; Li, M.; Stevens, T. Store-operated calcium entry promotes shape change in pulmonary endothelial cells expressing Trp1. Am. J. Physiol. 1998, 275, L574–L582. [Google Scholar] [CrossRef]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers-assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [Green Version]
- Pellegrin, S.; Mellor, H. Actin stress fibres. J. Cell Sci. 2007, 120, 3491–3499. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Zhang, J.J.; Huang, X.Y. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.K.; Lin, Y.H.; Chang, H.A.; Lai, Y.S.; Chen, Y.C.; Huang, S.C.; Chou, C.Y.; Chiu, W.T. Chemoresistant ovarian cancer enhances its migration abilities by increasing store-operated Ca2+ entry-mediated turnover of focal adhesions. J. Biomed. Sci. 2020, 27, 36. [Google Scholar] [CrossRef]
- Go, C.K.; Hooper, R.; Aronson, M.R.; Schultz, B.; Cangoz, T.; Nemani, N.; Zhang, Y.; Madesh, M.; Soboloff, J. The Ca2+ export pump PMCA clears near-membrane Ca2+ to facilitate store-operated Ca2+ entry and NFAT activation. Sci. Signal. 2019, 12, eaaw2627. [Google Scholar] [CrossRef]
- Wang, Y.; Kuramitsu, Y.; Ueno, T.; Suzuki, N.; Yoshino, S.; Iizuka, N.; Zhang, X.; Akada, J.; Oka, M.; Nakamura, K. Proteomic differential display identifies upregulated vinculin as a possible biomarker of pancreatic cancer. Oncol. Rep. 2012, 28, 1845–1850. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, J.; Xu, H.; Yang, Y.; Li, W.; Wu, H.; Liu, C. Extremely low frequency electromagnetic fields promote mesenchymal stem cell migration by increasing intracellular Ca2+ and activating the FAK/Rho GTPases signaling pathways in vitro. Stem. Cell Res. Ther. 2018, 9, 143. [Google Scholar] [CrossRef]
- Li, F.; Abuarab, N.; Sivaprasadarao, A. Reciprocal regulation of actin cytoskeleton remodelling and cell migration by Ca2+ and Zn2+: Role of TRPM2 channels. J. Cell Sci. 2016, 129, 2016–2029. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Guerrero, A.M.; Espinosa-Bermejo, N.; Sanchez-Lopez, I.; Macartney, T.; Pascual-Caro, C.; Orantos-Aguilera, Y.; Rodriguez-Ruiz, L.; Perez-Oliva, A.B.; Mulero, V.; Pozo-Guisado, E.; et al. RAC1-dependent ORAI1 translocation to the leading edge supports lamellipodia formation and directional persistence. Sci. Rep. 2020, 10, 6580. [Google Scholar] [CrossRef] [Green Version]
- Ballestrem, C.; Wehrle-Haller, B.; Hinz, B.; Imhof, B.A. Actin-dependent lamellipodia formation and microtubule-dependent tail retraction control-directed cell migration. Mol. Biol. Cell 2000, 11, 2999–3012. [Google Scholar] [CrossRef] [Green Version]
- Zabe, M.; Dean, W.L. Plasma membrane Ca (2+)-ATPase associates with the cytoskeleton in activated platelets through a PDZ-binding domain. J. Biol. Chem. 2001, 276, 14704–14709. [Google Scholar] [CrossRef] [Green Version]
- Dalghi, M.G.; Ferreira-Gomes, M.; Rossi, J.P. Regulation of the plasma membrane calcium ATPases by the actin cytoskeleton. Biochem. Biophys. Res. Commun. 2018, 506, 347–354. [Google Scholar] [CrossRef]
- Lee, S.; Kumar, S. Cofilin is required for polarization of tension in stress fiber networks during migration. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Redondo, P.C.; Harper, M.T.; Rosado, J.A.; Sage, S.O. A role for cofilin in the activation of store-operated calcium entry by de novo conformational coupling in human platelets. Blood 2006, 107, 973–979. [Google Scholar] [CrossRef]
- Pla, A.F.; Gkika, D. Emerging role of TRP channels in cell migration: From tumor vascularization to metastasis. Front. Physiol. 2013, 4, 311. [Google Scholar] [CrossRef] [Green Version]
- Ju, R.J.; Stehbens, S.J.; Haass, N.K. The role of melanoma cell-stroma interaction in cell motility, invasion, and metastasis. Front. Med. 2018, 5, 307. [Google Scholar] [CrossRef] [Green Version]
- Varga, K.; Hollósi, A.; Pászty, K.; Hegedűs, L.; Szakács, G.; Tímár, J.; Papp, B.; Enyedi, Á.; Padányi, R. Expression of calcium pumps is differentially regulated by histone deacetylase inhibitors and estrogen receptor alpha in breast cancer cells. BMC Cancer 2018, 18, 1029. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naffa, R.; Padányi, R.; Ignácz, A.; Hegyi, Z.; Jezsó, B.; Tóth, S.; Varga, K.; Homolya, L.; Hegedűs, L.; Schlett, K.; et al. The Plasma Membrane Ca2+ Pump PMCA4b Regulates Melanoma Cell Migration through Remodeling of the Actin Cytoskeleton. Cancers 2021, 13, 1354. https://doi.org/10.3390/cancers13061354
Naffa R, Padányi R, Ignácz A, Hegyi Z, Jezsó B, Tóth S, Varga K, Homolya L, Hegedűs L, Schlett K, et al. The Plasma Membrane Ca2+ Pump PMCA4b Regulates Melanoma Cell Migration through Remodeling of the Actin Cytoskeleton. Cancers. 2021; 13(6):1354. https://doi.org/10.3390/cancers13061354
Chicago/Turabian StyleNaffa, Randa, Rita Padányi, Attila Ignácz, Zoltán Hegyi, Bálint Jezsó, Sarolta Tóth, Karolina Varga, László Homolya, Luca Hegedűs, Katalin Schlett, and et al. 2021. "The Plasma Membrane Ca2+ Pump PMCA4b Regulates Melanoma Cell Migration through Remodeling of the Actin Cytoskeleton" Cancers 13, no. 6: 1354. https://doi.org/10.3390/cancers13061354






