G4-QuadScreen: A Computational Tool for Identifying Multi-Target-Directed Anticancer Leads against G-Quadruplex DNA
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Cancer
1.2. G-Quadruplex as a Potential Anticancer Target
1.3. Multi-Target Drug Designing
1.4. Multi-Target, Multi-Tasking QSAR Modeling
1.5. Aim of the Study
2. Materials and Methods
2.1. Dataset Collection and Curation
2.1.1. Dataset Collection
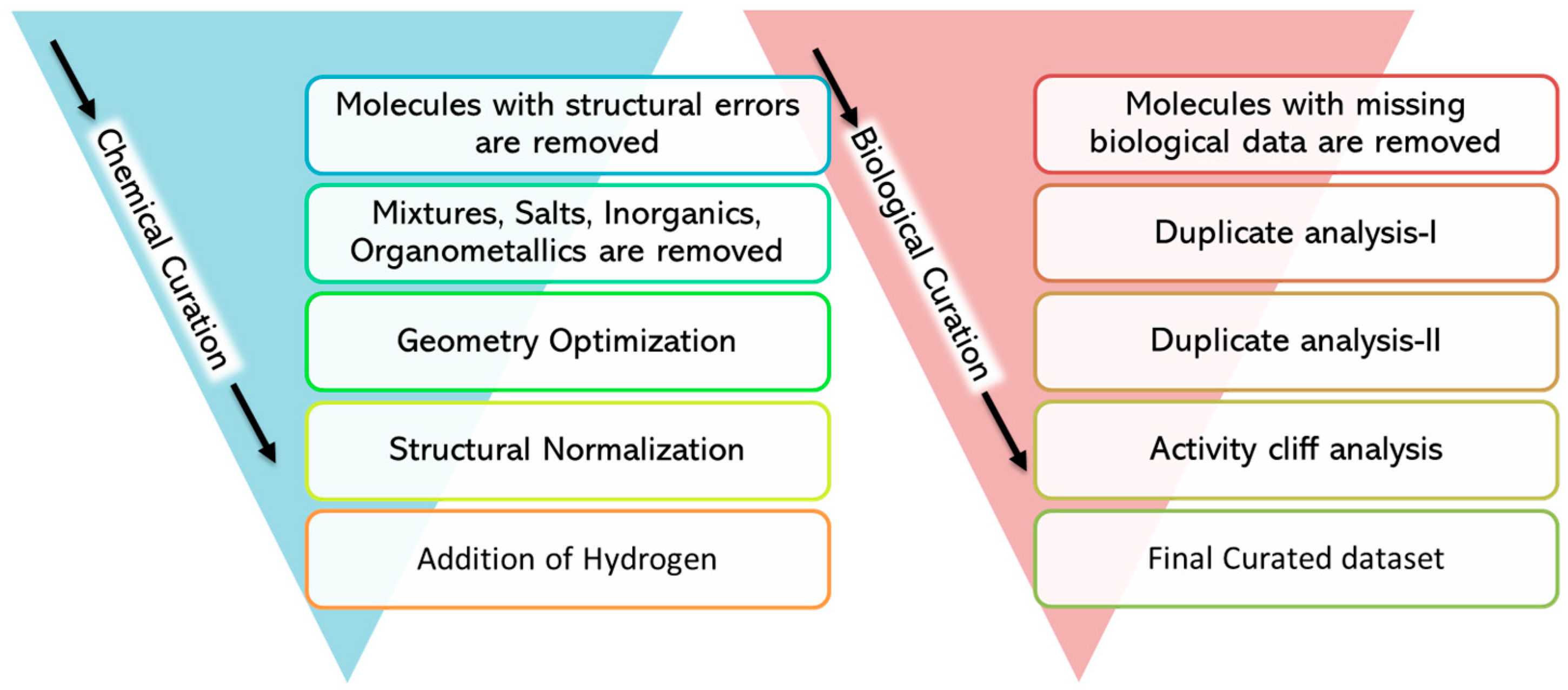
2.1.2. Data Curation
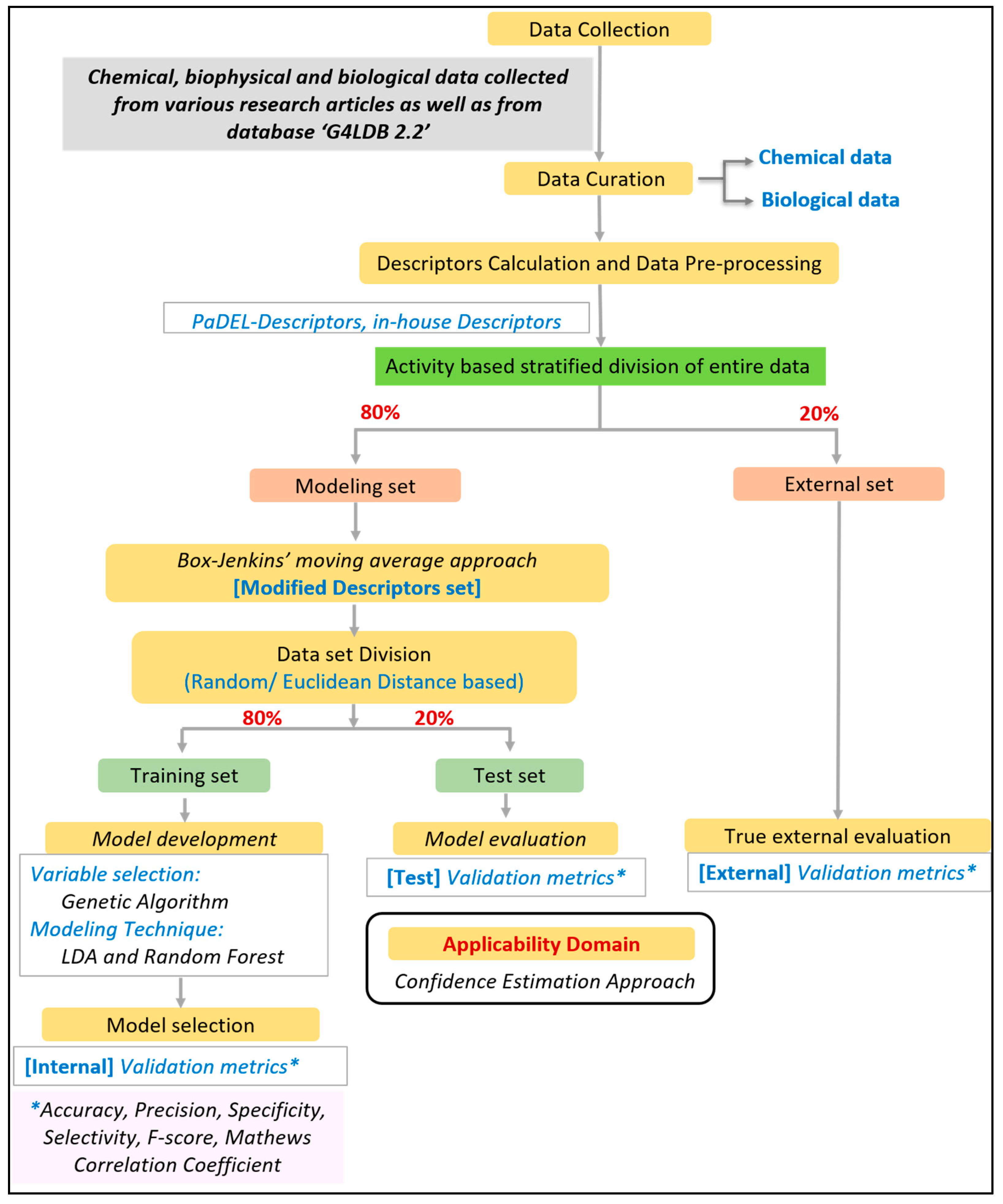
2.2. Descriptor Calculation and Data Preprocessing
2.3. Model Development and Validation
2.4. Applicability Domain
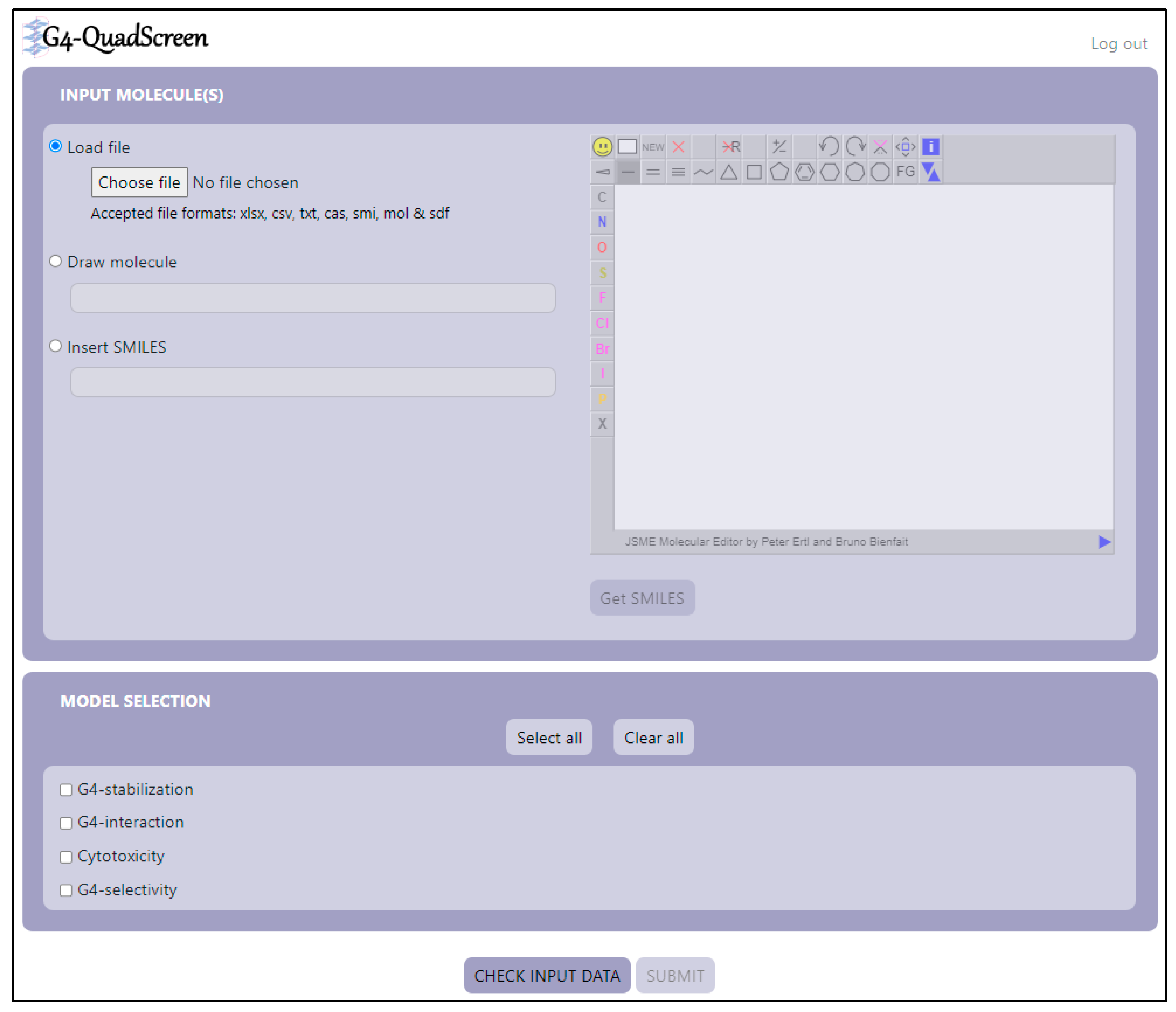
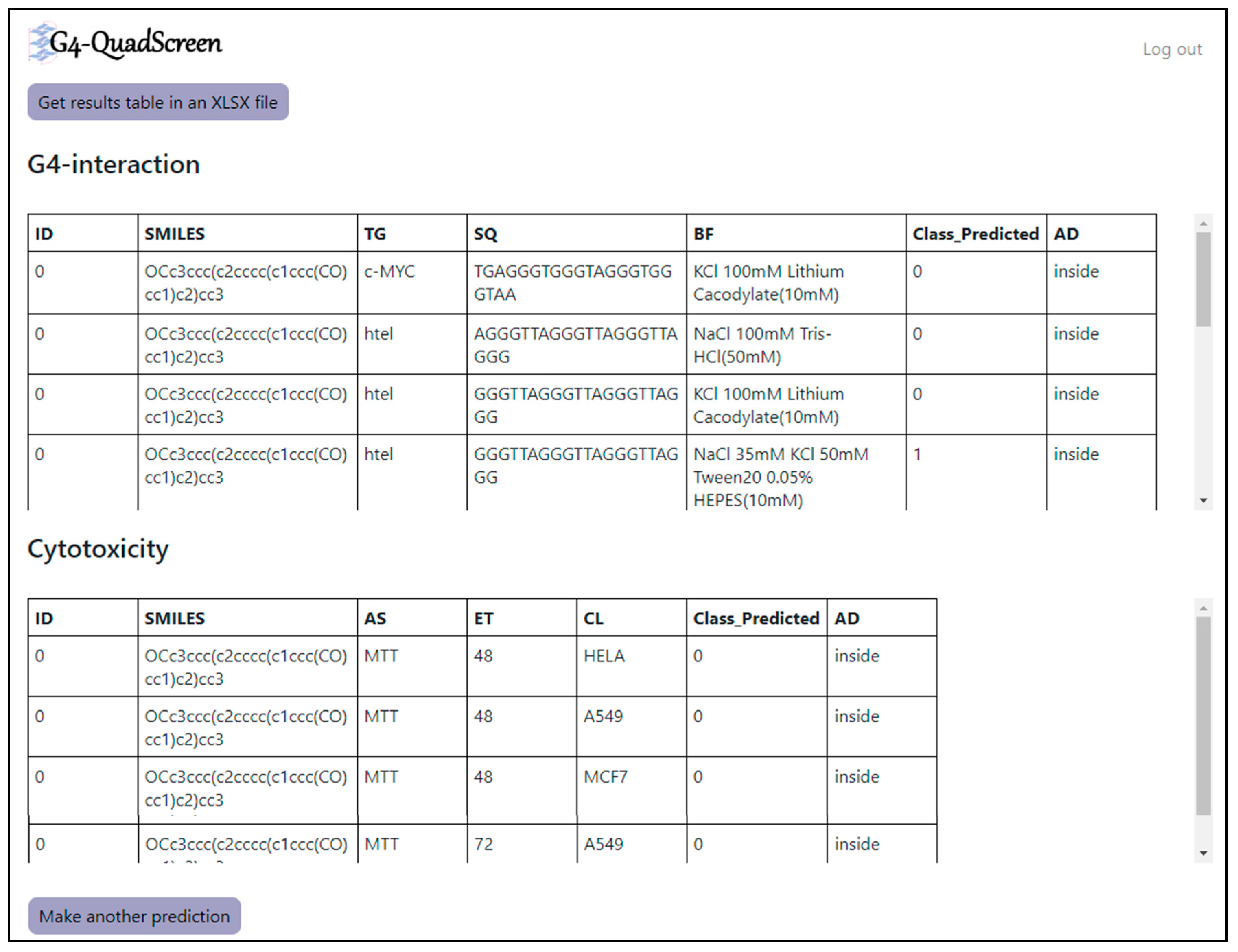
2.5. G4-QuadScreen Web-Based Computational Tool
2.6. Virtual Screening and Molecular Docking
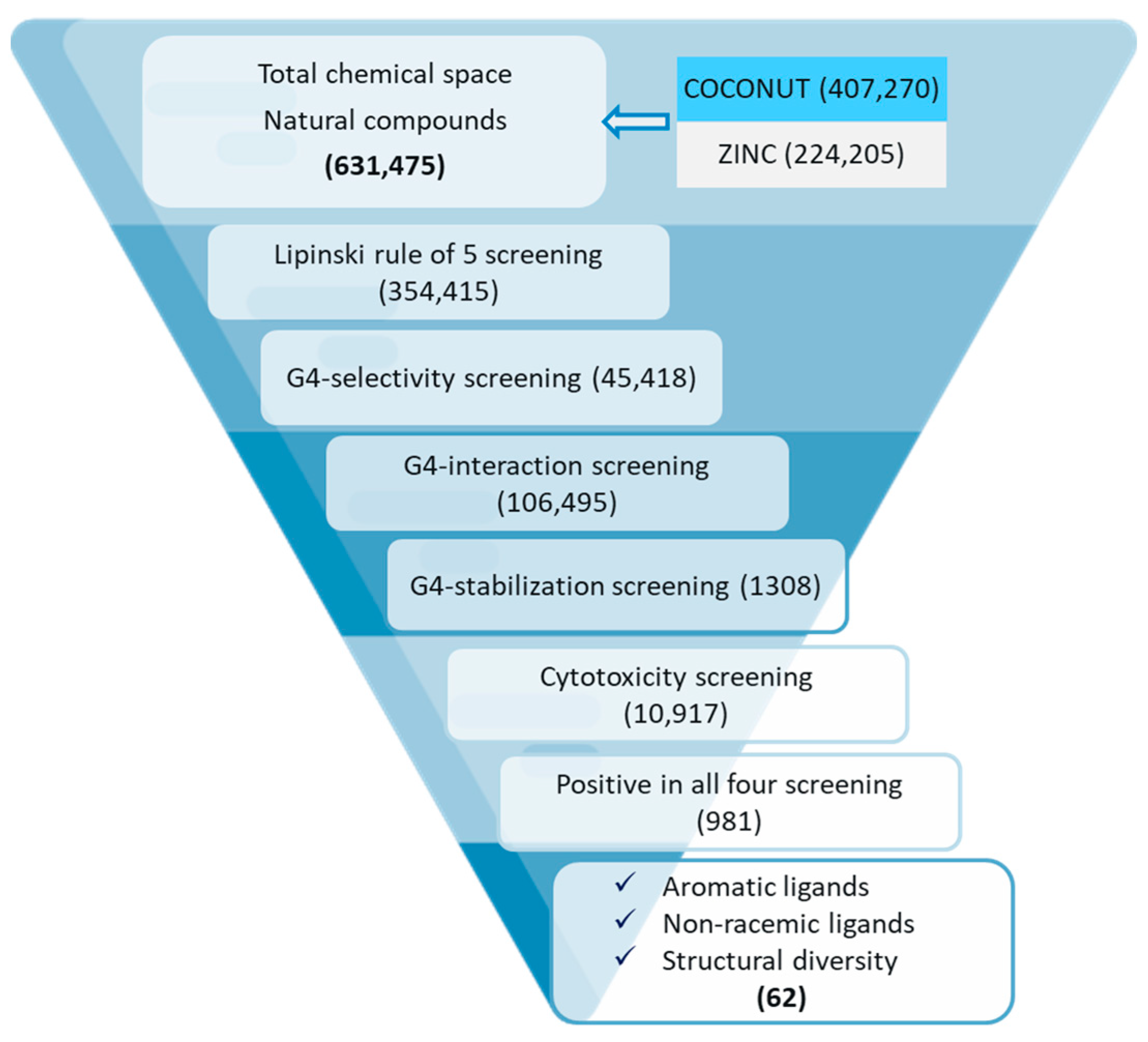
2.6.1. Virtual Screening
2.6.2. Molecular Docking
2.7. FRET Melting Experiments of Screened Compounds
2.8. TO Displacement (FID) Assay of Selected Compounds
2.9. Cell-Based Assays of Selective Compounds
2.9.1. Cell Culture
2.9.2. Cell Viability Assay
3. Results and Discussion
3.1. G4 Selectivity Model
3.2. G4 Interaction Model
3.3. G4 Stabilization Model
3.4. Cytotoxicity Model
3.5. G4-QuadScreen Web-Based Computational Tool
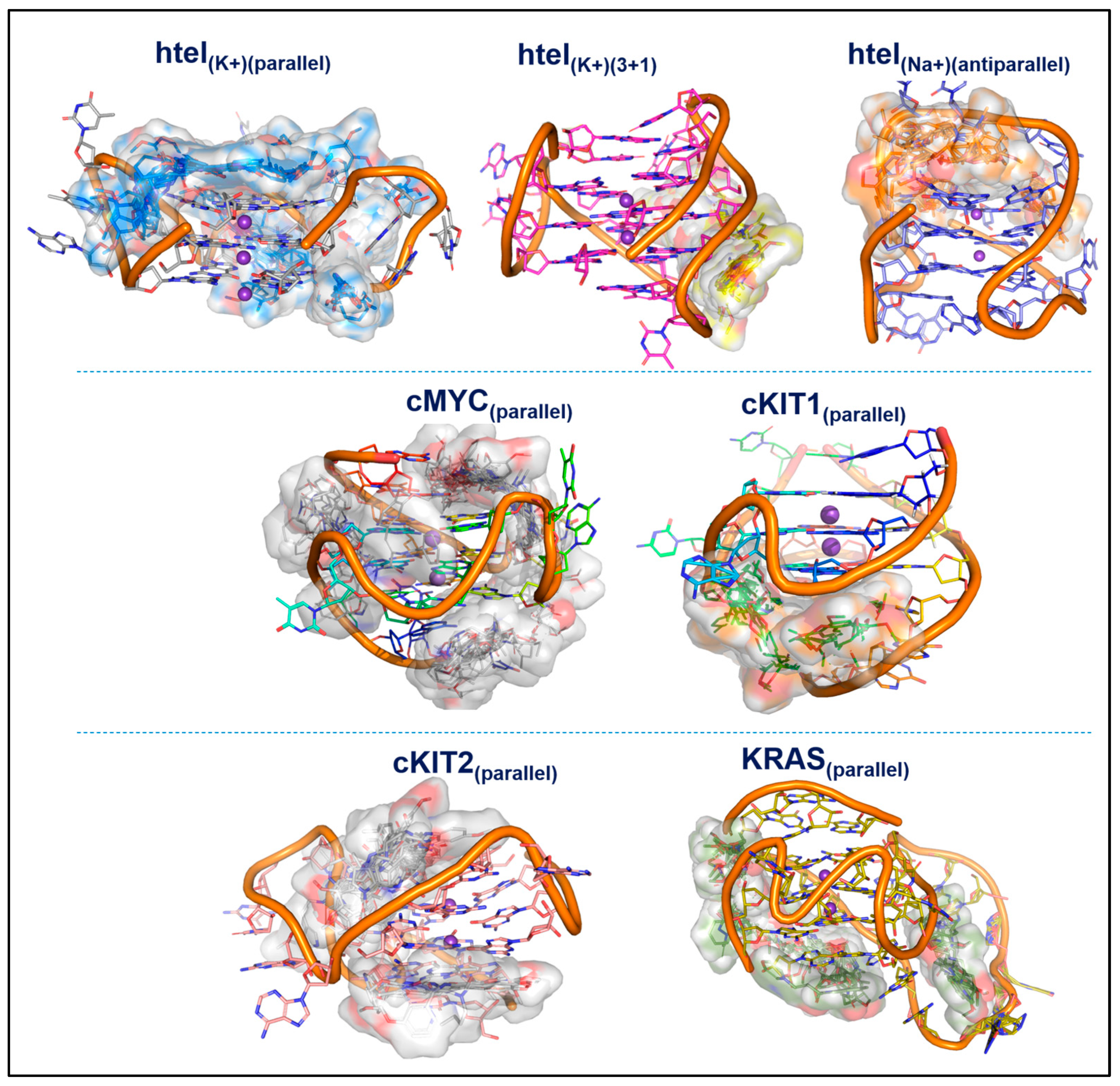
3.6. Molecular Docking
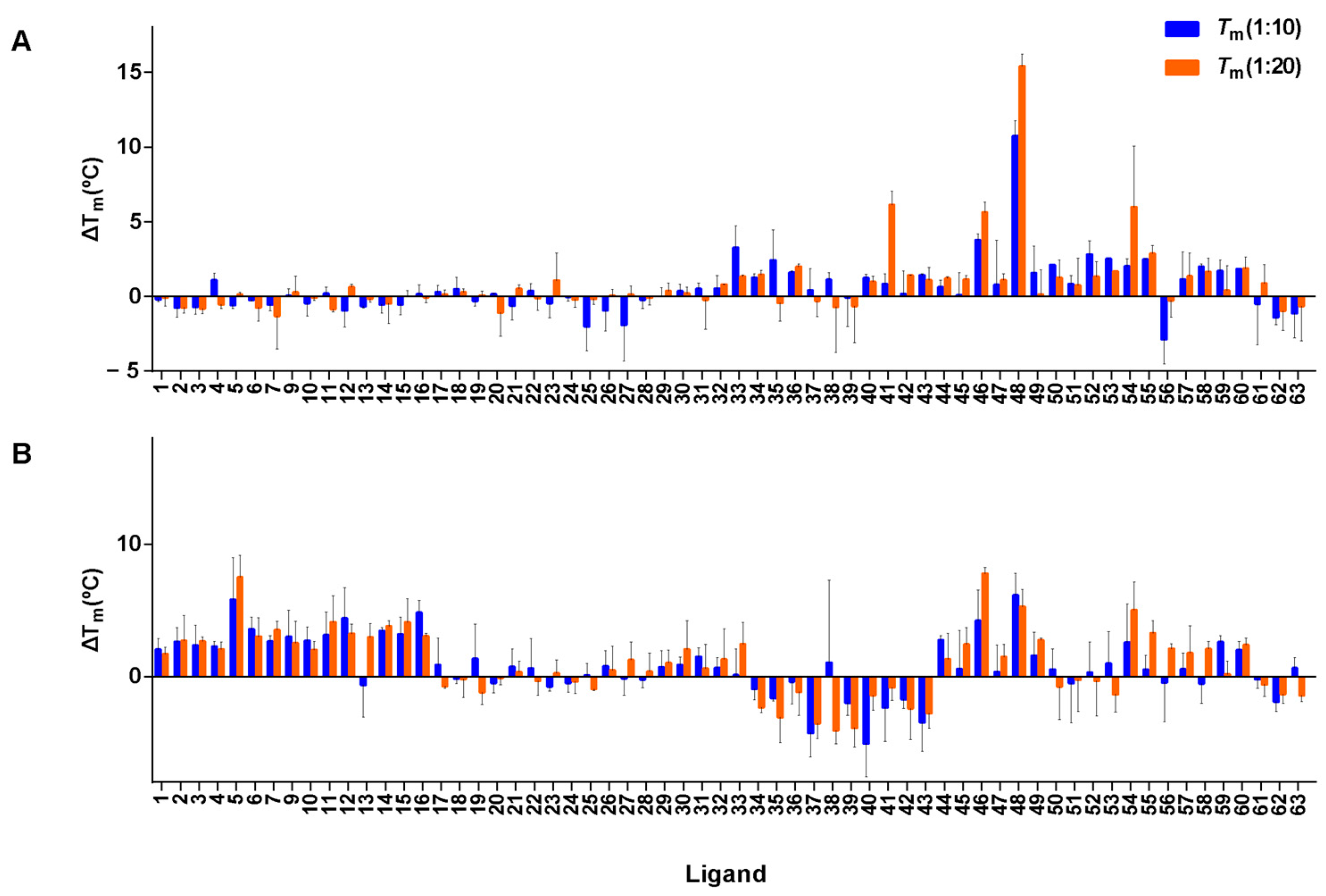
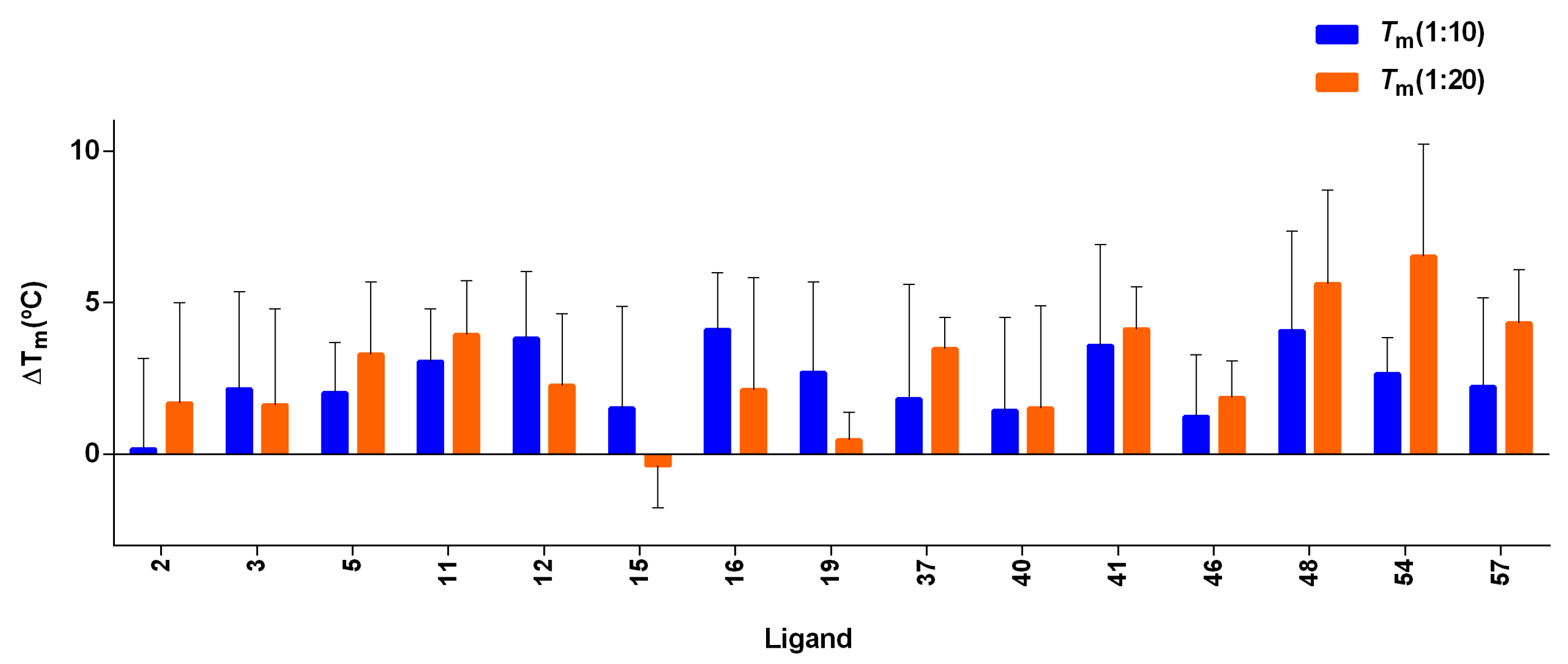
3.7. FRET Melting Experiments of Screened Compounds
3.8. FID-TO Displacement Assay of Selective Compounds
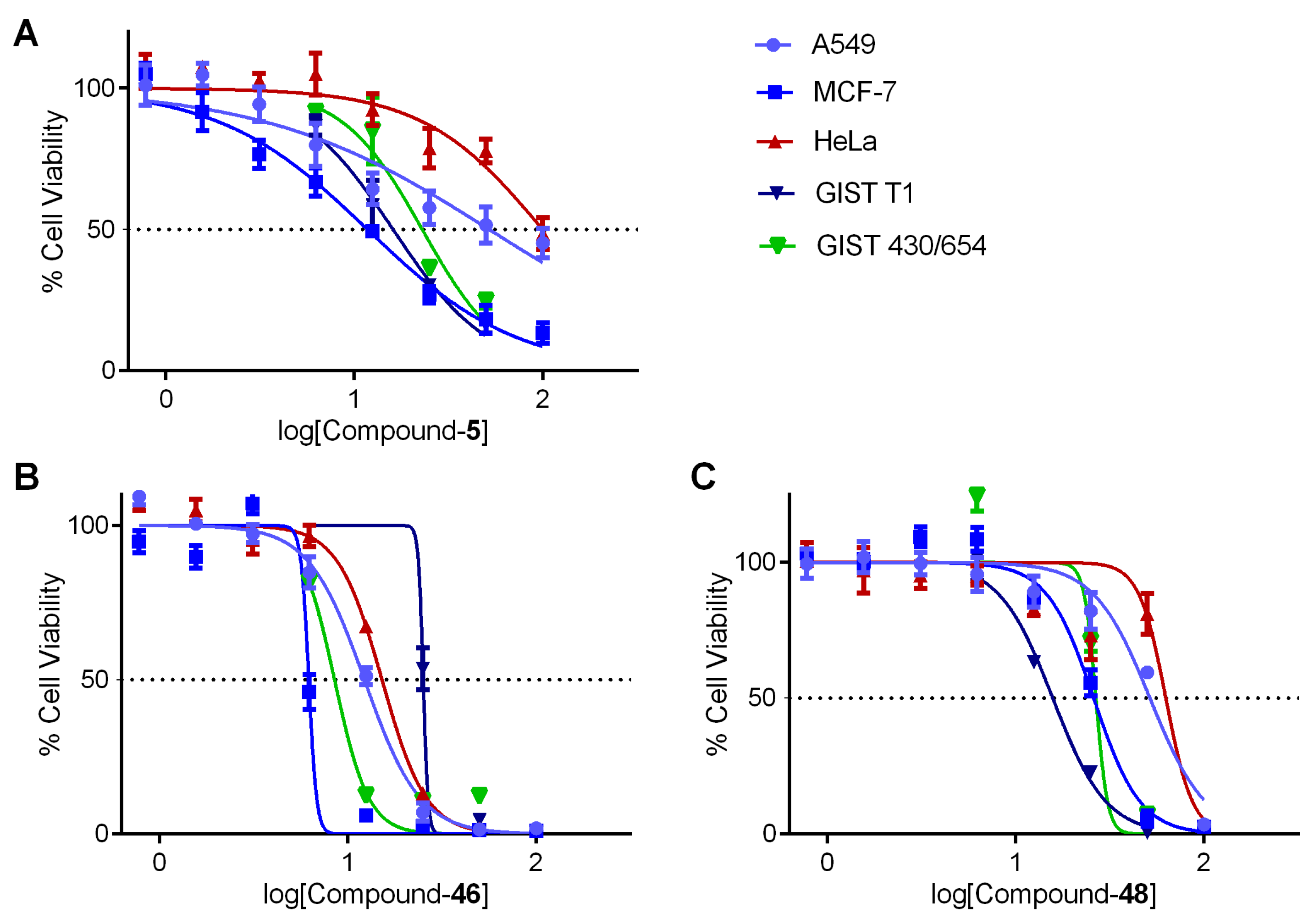
3.9. Cell-Based Assays of Selective Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Hoelder, S.; Clarke, P.A.; Workman, P. Discovery of small molecule cancer drugs: Successes, challenges and opportunities. Mol. Oncol. 2012, 6, 155–176. [Google Scholar] [CrossRef] [Green Version]
- Sawyers, C. Targeted cancer therapy. Nature 2004, 432, 294–297. [Google Scholar] [CrossRef]
- Neves, H.; Kwok, H.F. Recent advances in the field of anti-cancer immunotherapy. BBA Clin. 2015, 3, 280–288. [Google Scholar] [CrossRef] [Green Version]
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in Cancer Treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Neidle, S. Human Telomeric G-Quadruplex: The Current Status of Telomeric G-Quadruplexes as Therapeutic Targets in Human Cancer: G-Quadruplexes as Cancer Drug Targets. FEBS J. 2009, 277, 1118–1125. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Teulade-Fichou, M.-P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Cadoni, E.; De Paepe, L.; Manicardi, A.; Madder, A. Beyond small molecules: Targeting G-quadruplex structures with oligonucleotides and their analogues. Nucleic Acids Res. 2021, 49, 6638–6659. [Google Scholar] [CrossRef]
- Sun, Z.-Y.; Wang, X.-N.; Cheng, S.-Q.; Su, X.-X.; Ou, T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid—Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wu, Y.; Zhang, W. G-Quadruplex Structures and Their Interaction Diversity with Ligands. ChemMedChem 2014, 9, 899–911. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, M.P.; Morales, J.C.; Galan, M.C. Binding and Beyond: What Else Can G-Quadruplex Ligands Do? Eur. J. Org. Chem. 2019, 2019, 4995–5017. [Google Scholar] [CrossRef]
- Taetz, S.; Baldes, C.; Mürdter, T.E.; Kleideiter, E.; Piotrowska, K.; Bock, U.; Haltner-Ukomadu, E.; Mueller, J.; Huwer, H.; Schaefer, U.F.; et al. Biopharmaceutical Characterization of the Telomerase Inhibitor BRACO19. Pharm. Res. 2006, 23, 1031–1037. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Popovic-Nikolicb, M.R.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Baginski, M.; Serbakowska, K. In silico design of telomerase inhibitors. Drug Discov. Today 2020, 25, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Yazdani, K.K.; Jordan, D.; Yang, M.; Fullenkamp, C.R.; Calabrese, D.R.; Boer, R.; Hilimire, T.; Allen, T.E.H.; Khan, R.T.; Schneekloth, J.S., Jr. Machine Learning Informs RNA-Binding Chemical Space. Angew. Chem. Int. Ed. Engl. 2023, 135, e202211358. [Google Scholar] [CrossRef]
- Disney, M.D.; Winkelsas, A.M.; Velagapudi, S.P.; Southern, M.; Fallahi, M.; Childs-Disney, J.L. Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs. ACS Chem. Biol. 2016, 11, 1720–1728. [Google Scholar] [CrossRef] [Green Version]
- Casañola-Martin, G.M.; Le-Thi-Thu, H.; Pérez-Giménez, F.; Marrero-Ponce, Y.; Merino-Sanjuán, M.; Abad, C.; González-Díaz, H. Multi-output model with Box–Jenkins operators of linear indices to predict multi-target inhibitors of ubiquitin-proteasome pathway. Mol. Divers. 2015, 19, 347–356. [Google Scholar] [CrossRef]
- Hill, S.A. Statistics. In Foundations of Anesthesia; Elsevier: Amsterdam, The Netherlands, 2006; pp. 207–217. ISBN 978-0-323-03707-5. [Google Scholar]
- Li, Q.; Xiang, J.-F.; Yang, Q.-F.; Sun, H.-X.; Guan, A.-J.; Tang, Y.-L. G4LDB: A database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013, 41, D1115–D1123. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-H.; Yang, Q.-F.; Lin, X.; Chen, D.; Wang, Z.-Y.; Chen, B.; Han, H.-Y.; Chen, H.-D.; Cai, K.-C.; Li, Q.; et al. G4LDB 2.2: A database for discovering and studying G-quadruplex and i-Motif ligands. Nucleic Acids Res. 2022, 50, D150–D160. [Google Scholar] [CrossRef] [PubMed]
- Ambure, P.; Cordeiro, M.N.D.S. Importance of Data Curation in QSAR Studies Especially While Modeling Large-Size Datasets. In Ecotoxicological QSARs; Roy, K., Ed.; Methods in Pharmacology and Toxicology; Springer: New York, NY, USA, 2020; pp. 97–109. ISBN 978-1-07-160149-5. [Google Scholar]
- Fourches, D.; Muratov, E.; Tropsha, A. Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model. 2016, 56, 1243–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambure, P.; Bhat, J.; Puzyn, T.; Roy, K. Identifying natural compounds as multi-target-directed ligands against Alzheimer’s disease: An in silico approach. J. Biomol. Struct. Dyn. 2019, 37, 1282–1306. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2010, 32, 1466–1474. [Google Scholar] [CrossRef]
- Ambure, P.; Aher, R.B.; Gajewicz, A.; Puzyn, T.; Roy, K. “NanoBRIDGES” software: Open access tools to perform QSAR and nano-QSAR modeling. Chemom. Intell. Lab. Syst. 2015, 147, 1–13. [Google Scholar] [CrossRef]
- Ambure, P.; Halder, A.K.; Díaz, H.G.; Cordeiro, M.N.D.D.S. QSAR-Co: An Open Source Software for Developing Robust Multitasking or Multitarget Classification-Based QSAR Models. J. Chem. Inf. Model. 2019, 59, 2538–2544. [Google Scholar] [CrossRef]
- Davies, O.L.; Snedecor, G.W.; Cochran, W.G. Statistical Methods, 8th ed.; The Iowa State University Press: Ames, IA, USA, 1989; ISBN 978-0813815619. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wilks, S.S. Certain generalizations in the analysis of variance. Biometrika 1932, 24, 471–494. [Google Scholar] [CrossRef]
- Langevin, M.; Grebner, C.; Güssregen, S.; Sauer, S.; Li, Y.; Matter, H.; Bianciotto, M. Impact of Applicability Domains to Generative Artificial Intelligence. ACS Omega 2023, 8, 23148–23167. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, J.; Jeliazkova, N.; Aldenberg, T. QSAR Applicability Domain Estimation by Projection of the Training Set in Descriptor Space: A Review. Altern. Lab. Anim. 2005, 33, 445–459. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, H.; Li, C.; Zhang, J.Z.H.; Ji, C. MolGpka: A Web Server for Small Molecule pKa Prediction Using a Graph-Convolutional Neural Network. J. Chem. Inf. Model. 2021, 61, 3159–3165. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [Green Version]
- Largy, E.; Hamon, F.; Teulade-Fichou, M.-P. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal. Bioanal. Chem. 2011, 400, 3419–3427. [Google Scholar] [CrossRef]
- Serrano, C.; Mariño-Enríquez, A.; Tao, D.L.; Ketzer, J.; Eilers, G.; Zhu, M.; Yu, C.; Mannan, A.M.; Rubin, B.P.; Demetri, G.D.; et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br. J. Cancer 2019, 120, 612–620. [Google Scholar] [CrossRef] [Green Version]
- Bauer, S.; Duensing, A.; Demetri, G.D.; Fletcher, J.A. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene 2007, 26, 7560–7568. [Google Scholar] [CrossRef] [Green Version]
- Paul, R.; Das, T.; Debnath, M.; Chauhan, A.; Dash, J. G-Quadruplex-Binding Small Molecule Induces Synthetic Lethality in Breast Cancer Cells by Inhibiting c-MYC and BCL2 Expression. Chembiochem 2020, 21, 963–970. [Google Scholar] [CrossRef] [PubMed]
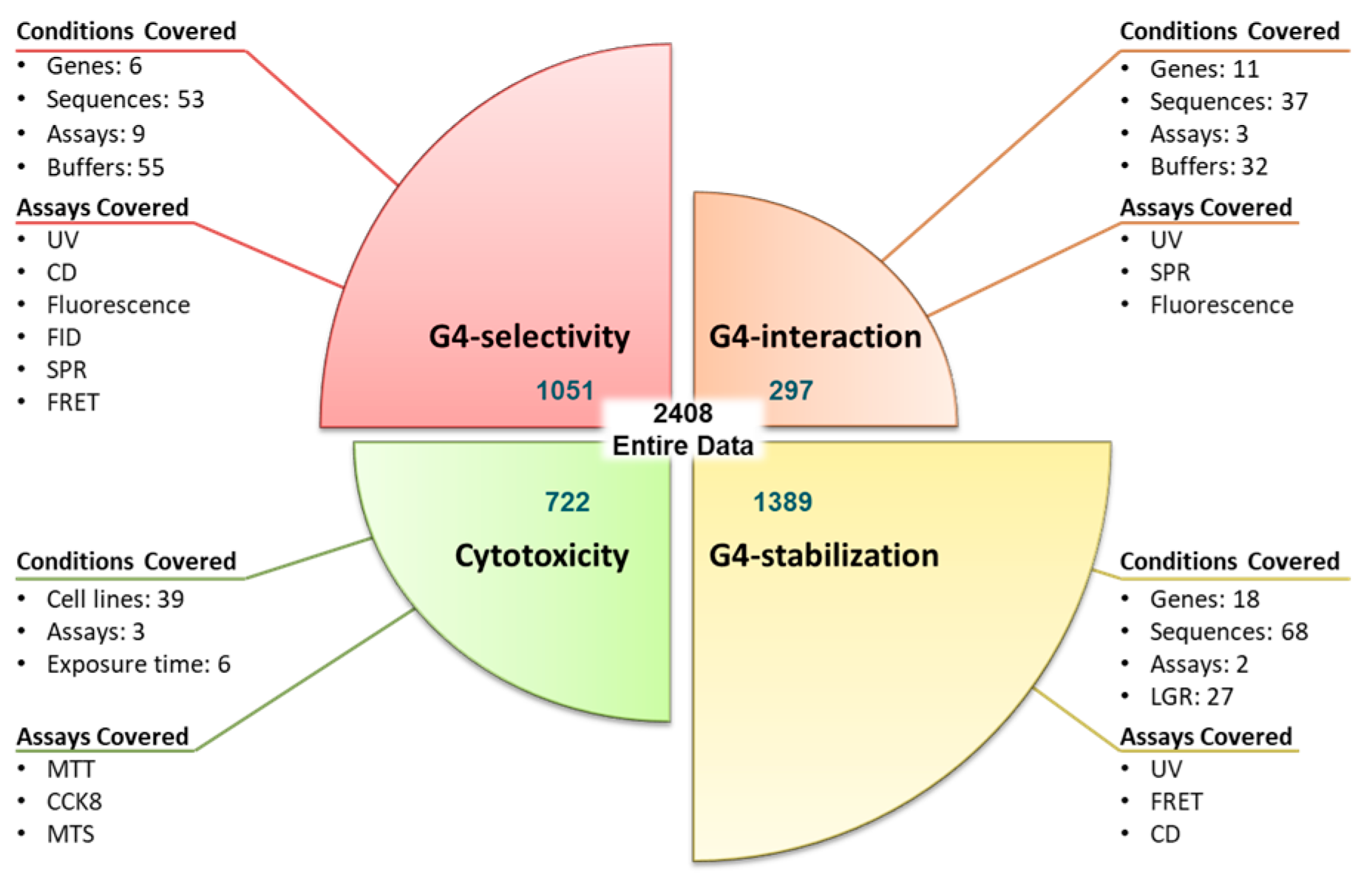

| Gene | Function | Cancer Type |
|---|---|---|
| cMYC | Cell proliferation, differentiation, and apoptosis (PDA) | Cervical carcinoma, myeloid leukemia (MyL) |
| cKIT | Cell PDA, motility, adhesion, angiogenesis | Gastrointestinal cancer, MyL |
| KRAS | Cell PDA | Lung, pancreatic cancer, MyL |
| BCL2 | Oncogenesis through cell death resistance | Small-cell lung cancer, breast cancer |
| hTel | Cell cycle arrest and cell death | Numerous types of cancer |
| Model | Experiment | Outcome |
|---|---|---|
| G4 interaction | UV, SPR, fluorescence titrations | The absorption and fluorescence response of the molecules upon binding with G4s and duplex structures allow us to determine binding affinity constants, thus providing information about interaction capacity and the selectivity of ligands towards G4. |
| G4 stabilization | UV, FRET, CD melting data | ∆Tm (change in melting temperature) quantitatively determines the stabilization effect of ligands over the secondary structure of DNA. Thus, the G4 stabilization effect and G4 selectivity can be evaluated. |
| G4 selectivity | Combination of above experiments | When the interaction capacity and stabilization effect are one order higher in G4 than that for duplex DNA, the ligand is considered selective. |
| Cytotoxicity | MTT, CCK8, MTS | These assays provide IC50 values of ligands against various cancer cell lines, thus providing information on the activity of ligands at a cellular level. |
| Model | Threshold |
|---|---|
| G4 selectivity | When ΔTm and Kd values are higher in G4 than those in duplex DNA, the ligand is considered selective, or else non-selective. |
| G4 interaction | Positive: Kd < 1 µM Negative: Kd ≥ 1 µM |
| G4 stabilization | Positive: ΔTm ≥ 15 °C Negative ΔTm < 15 °C |
| Cytotoxicity | Positive: IC50 < 10 µM Negative: IC50 ≥ 10 µM |
| Model Category | Parameters | Train | Train 10-Fold CV | Test | Ext | Ext in-AD |
|---|---|---|---|---|---|---|
| I. G4 Selectivity Descriptor: 15 Random forest | No. of Compounds | P: 493 N: 175 Total: 668 | P: 493 N: 175 Total: 668 | P: 127 N: 39 Total: 166 | P: 159 N: 58 Total: 217 | P: 156 N: 50 Total: 206 |
| Accuracy% | 99.850 | 81.280 ± 5.250 | 86.747 | 90.320 | 91.260 | |
| Precision% | 99.800 | 83.860 ± 4.600 | 90.700 | 92.590 | 92.590 | |
| Sensitivity% | 100 | 92.890 ± 4.600 | 92.130 | 94.340 | 96.150 | |
| Specificity% | 99.430 | 48.370 ± 17.250 | 69.230 | 79.310 | 76.000 | |
| F-measure | 0.9999 | 0.880 ± 0.035 | 0.914 | 0.935 | 0.943 | |
| MCC | 0.996 | 0.485 ± 0.158 | 0.625 | 0.749 | 0.755 |
| Model Category | Parameters | Train | Train 10-Fold CV | Test | Ext | Ext in-AD |
|---|---|---|---|---|---|---|
| II. G4 Interaction (Kd) Descriptor: 10 LDA | No. of Compounds | P: 100 N: 95 Total: 195 | P: 100 N: 95 Total: 195 | P: 21 N: 27 Total: 48 | P: 29 N: 25 Total: 54 | P: 23 N: 16 Total: 39 |
| Accuracy% | 92.821 | 86.340 ± 9.734 | 85.417 | 83.330 | 89.740 | |
| Precision% | 93.000 | 84.014 ± 11.390 | 75.000 | 88.460 | 95.240 | |
| Sensitivity% | 93.000 | 91.999 ± 7.483 | 100.000 | 79.310 | 86.960 | |
| Specificity% | 92.632 | 80.670 ± 14.942 | 74.074 | 88.000 | 93.750 | |
| F-measure | 0.930 | 0.875 ± 8.660 | 0.857 | 0.836 | 0.909 | |
| MCC | 0.856 | 0.736 ± 18.770 | 0.745 | 0.672 | 0.796 |
| Model Category | Parameters | Train | Train 10-Fold CV | Test | Ext | Ext in-AD |
|---|---|---|---|---|---|---|
| III. G4 Stabilization (ΔTm) Threshold Positive: ΔTm ≥ 15 °C Negative: ΔTm < 15 °C Descriptor: 10 Random Forest | No. of Compounds | P: 498 N: 394 Total: 892 | P: 498 N: 394 Total: 892 | P: 129 N: 93 Total: 222 | P: 154 N: 121 Total: 275 | P: 119 N: 85 Total: 204 |
| Accuracy% | 99.890 | 85.540 ± 4.970 | 90.540 | 85.091 | 85.440 | |
| Precision% | 100.000 | 86.990 ± 7.170 | 92.860 | 88.970 | 88.650 | |
| Sensitivity% | 99.800 | 88.335 ± 7.790 | 90.700 | 83.770 | 85.030 | |
| Specificity% | 100.000 | 81.885 ± 11.860 | 90.320 | 86.780 | 85.960 | |
| F-measure | 0.999 | 0.872 ± 0.044 | 0.918 | 0.863 | 0.868 | |
| MCC | 0.998 | 0.714 ± 0.098 | 0.807 | 0.701 | 0.707 |
| Model Category | Parameters | Train | Train 10-Fold CV | Test | Ext | Ext in-AD |
|---|---|---|---|---|---|---|
| IV. Cytotoxicity (MTT, MTS, CCK8) Threshold Positive: IC50 < 10 µM Negative: IC50 ≥ 10 µM Descriptor: 9 Random Forest | No. of Compounds | P: 244 N: 219 Total: 463 | P: 244 N: 219 Total: 463 | P: 71 N: 44 Total: 115 | P: 79 N: 65 Total: 144 | P: 74 N: 65 Total: 139 |
| Accuracy% | 100.000 | 69.740 ± 6.520 | 84.348 | 83.330 | 82.730 | |
| Precision% | 100.000 | 74.285 ± 12.010 | 92.060 | 85.710 | 83.560 | |
| Sensitivity% | 100.000 | 71.000 ± 18.620 | 81.690 | 83.540 | 83.560 | |
| Specificity% | 100.000 | 68.440 ± 20.270 | 88.640 | 83.080 | 81.820 | |
| F-measure | 1.000 | 0.870 ± 0.088 | 0.866 | 0.846 | 0.836 | |
| MCC | 1.000 | 0.426 ± 0.144 | 0.687 | 0.665 | 0.654 |
| G4 Model | Screening Criteria | Rationale |
|---|---|---|
| Selectivity | G4 Sequences: hTel: AGGGTTAGGGTTAGGGTTAGGG hTel: GGGTTAGGGTTAGGGTTAGGG cMYC: TGAGGGTGGGTAGGGTGGGTAA cKIT1: GGGAGGGCGCTGGGAGGAGGG cKIT2: GGGCGGGCGCGAGGGAGGGG KRAS: AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | ~50% of the modeling data comprise these sequences. |
| Buffer: KCl 10 mM LiCl 90 mM Lithium Cacodylate (10 mM) NaCl 100 mM Lithium Cacodylate (10 mM) KCl 100 mM Lithium Cacodylate (10 mM) | Frequently adopted in FRET experiments, and cover parallel, antiparallel, and hybrid topology of G4. | |
| Assay: FRET melting | ~43% of the modeling data are derived from FRET experiments. | |
| Interaction | G4 Sequences: hTel: AGGGTTAGGGTTAGGGTTAGGG hTel: GGGTTAGGGTTAGGGTTAGGG cMYC: TGAGGGTGGGTAGGGTGGGTAA cKIT1: GGGAGGGCGCTGGGAGGAGGG cKIT2: GGGCGGGCGCGAGGGAGGGG KRAS: AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | ~50% of the modeling data comprise these sequences. |
| Buffer: KCl 100 mM Lithium Cacodylate (10 mM) NaCl 35 mM KCl 50 mM Tween20 0.05% HEPES (10 mM) NaCl 100 mM Tris-HCl (50 mM) KCl 100 mM Tris-HCl (10 mM) | Frequently adopted in G4-Kd studies, and cover parallel, antiparallel, and hybrid topology of G4. | |
| Stabilization | G4 Sequences: hTel: AGGGTTAGGGTTAGGGTTAGGG hTel: GGGTTAGGGTTAGGGTTAGGG cMYC: TGAGGGTGGGTAGGGTGGGTAA cKIT1: GGGAGGGCGCTGGGAGGAGGG cKIT2: GGGCGGGCGCGAGGGAGGGG KRAS: AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | ~55% of the modeling data comprise these sequences. |
| Buffer: KCl 10 mM LiCl 90 mM Lithium Cacodylate (10 mM) NaCl 10 mM LiCl 90 mM Lithium Cacodylate (10 mM) | Frequently adopted in FRET experiments, and cover parallel, antiparallel, and hybrid topology of G4. | |
| Assay: FRET melting experiments | ~88% of the modeling data comprise this assay condition. | |
| Ligand to G4 ratio (LGR): 5, 10 | ~53% of the modeling data were obtained for these LGRs, and they have well-balanced class distribution. | |
| Cytotoxicity | Cell lines: HELA, A549, MCF7, A375, HCT116 | ~48% of the modeling data have the cell line condition of these cell lines. |
| Exposure time: 48, 72 h | ~95% of the modeling data have these exposure time conditions. | |
| Assay: MTT | ~90% of the modeling data are derived from the MTT assay. |
| G4 Motif | Average Binding Energies (kcal/mol) | G4 Pocket |
|---|---|---|
| hTel (antiparallel) | 9.11 | 5′end, Groove |
| hTel (3+1 hybrid) | 7.63 | Groove |
| hTel (parallel) | 7.63 | 3′end, Groove |
| cMYC | 7.83 | 5′end, 3′end, Groove |
| cKIT1 | 8.41 | 3′end, Groove |
| cKIT2 | 7.46 | 5′end, 3′end, Groove |
| KRAS | 7.77 | 5′end, 3′end, Groove |
| Compounds | IC50 (µM) | ||||
|---|---|---|---|---|---|
| A549 a | MCF-7 a | HeLa a | GIST T1 b | GIST 430/650 b | |
| 5 | >100 | 11.8 | >100 | 16.01 | 22.94 |
| 46 | 12.24 | 6.02 | 15.17 | 25.11 | 8.495 |
| 48 | 51.58 | 25.95 | 62.64 | 15.74 | 26.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat-Ambure, J.; Ambure, P.; Serrano-Candelas, E.; Galiana-Roselló, C.; Gil-Martínez, A.; Guerrero, M.; Martin, M.; González-García, J.; García-España, E.; Gozalbes, R. G4-QuadScreen: A Computational Tool for Identifying Multi-Target-Directed Anticancer Leads against G-Quadruplex DNA. Cancers 2023, 15, 3817. https://doi.org/10.3390/cancers15153817
Bhat-Ambure J, Ambure P, Serrano-Candelas E, Galiana-Roselló C, Gil-Martínez A, Guerrero M, Martin M, González-García J, García-España E, Gozalbes R. G4-QuadScreen: A Computational Tool for Identifying Multi-Target-Directed Anticancer Leads against G-Quadruplex DNA. Cancers. 2023; 15(15):3817. https://doi.org/10.3390/cancers15153817
Chicago/Turabian StyleBhat-Ambure, Jyotsna, Pravin Ambure, Eva Serrano-Candelas, Cristina Galiana-Roselló, Ariadna Gil-Martínez, Mario Guerrero, Margarita Martin, Jorge González-García, Enrique García-España, and Rafael Gozalbes. 2023. "G4-QuadScreen: A Computational Tool for Identifying Multi-Target-Directed Anticancer Leads against G-Quadruplex DNA" Cancers 15, no. 15: 3817. https://doi.org/10.3390/cancers15153817





