The β-Secretase 1 Enzyme as a Novel Therapeutic Target for Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
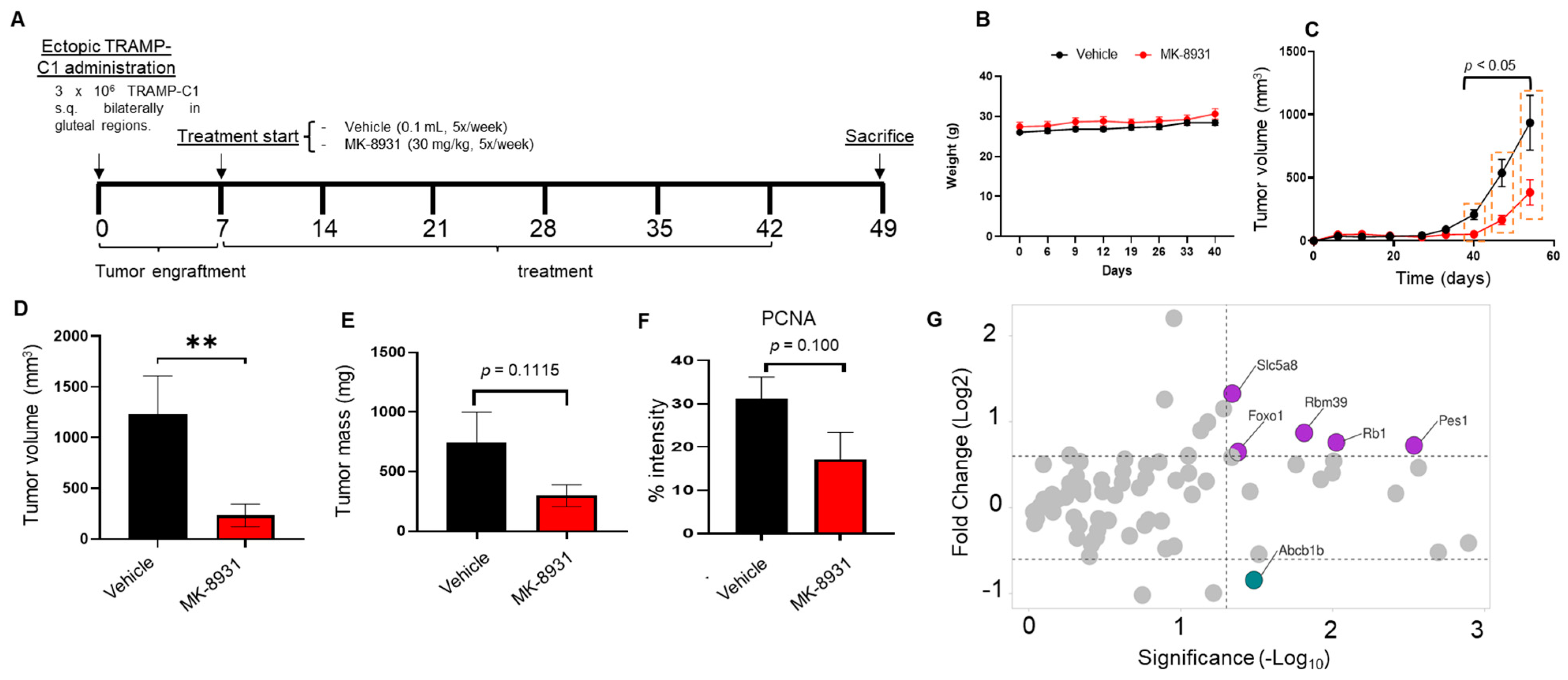
2.2. Animal Experiment
2.3. Archived Tissues and Immunohistochemistry (IHC)
2.4. Immunofluorescence (IF)
2.5. PET Imaging
2.6. Gene Expression
2.7. Pathway Specific PCR Array
2.8. BACE1 Activity Assay
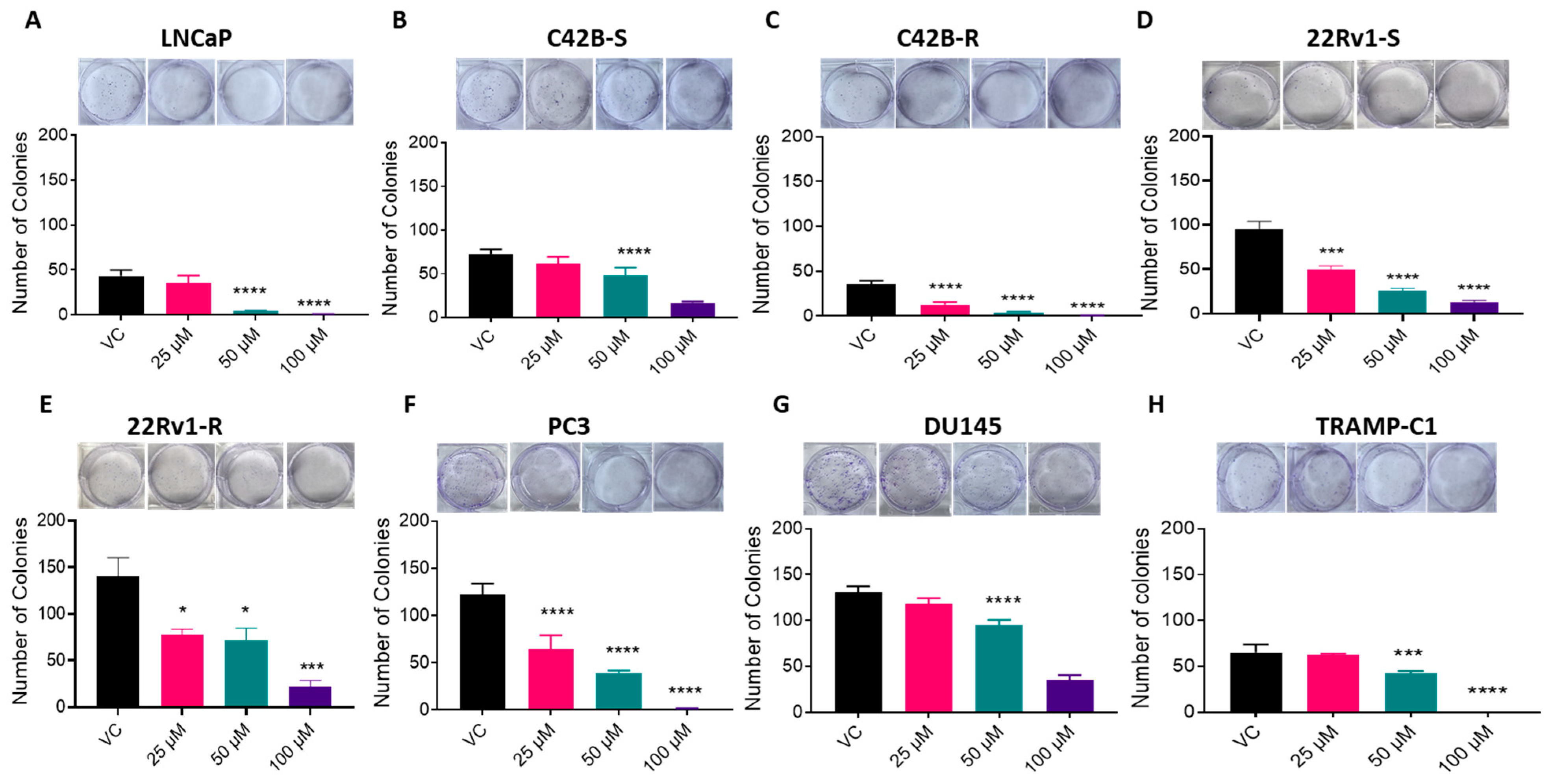
2.9. Colony Formation Assay
2.10. MTT Assay
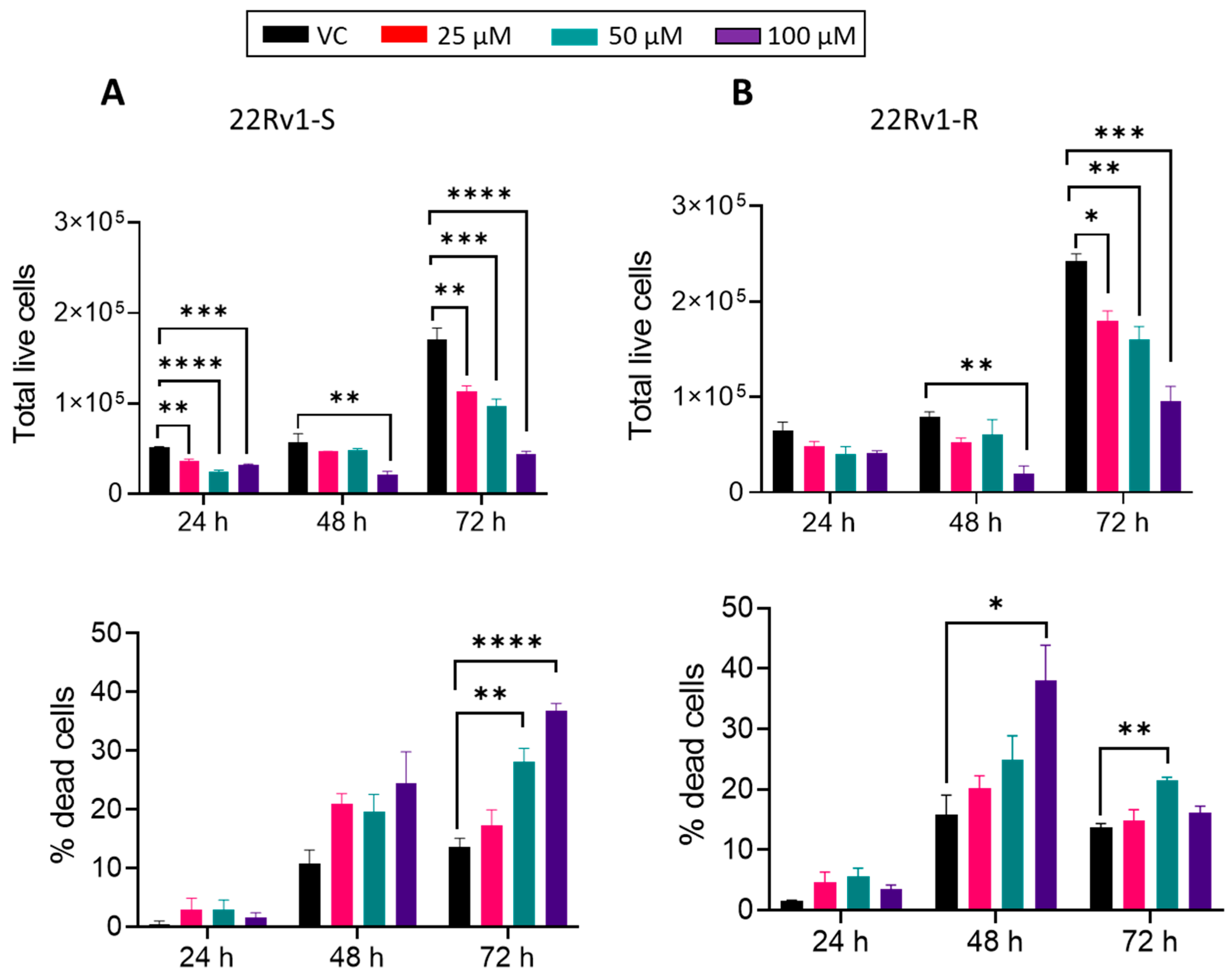
2.11. Trypan Blue Assay
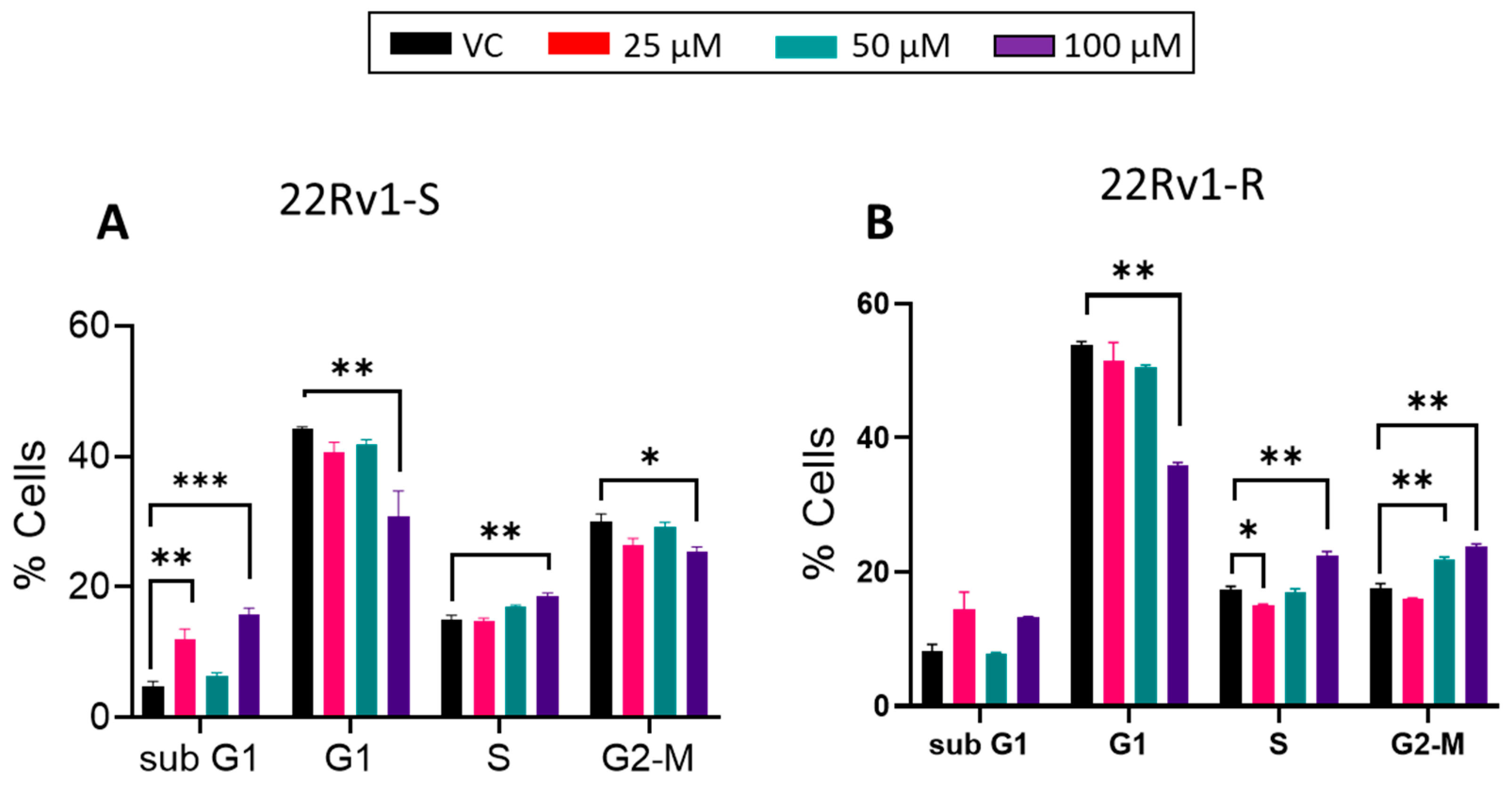
2.12. Cell Cycle Analysis
2.13. Statistical Analysis
3. Results
3.1. BACE1 Expression in PCa
3.2. BACE1 Gene Expression and Activity in PCa Cells
3.3. BACE1 Inhibition Causes Reduced Cell Viability in PCa Cells
3.4. Effect of BACE1 Inhibition on Cell Growth, Death, and Cell Cycle in Enzalutamide-Sensitive and -Resistant PCa Cells
3.5. BACE1 Inhibitor Reduces the Growth of PCa Allografts in Mice without Any Toxicity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Key Statistics for Prostate Cancer. Available online: https://www.cancer.org/cancer/types/prostate-cancer/about/key-statistics.html (accessed on 17 July 2022).
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. Ca Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Corn, P.G.; Thompson, T.C. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32, 5501–5511. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Alabi, B.R.; Liu, S.; Stoyanova, T. Current and emerging therapies for neuroendocrine prostate cancer. Pharmacol. Ther. 2022, 238, 108255. [Google Scholar] [CrossRef] [PubMed]
- Chow, V.W.; Mattson, M.P.; Wong, P.C.; Gleichmann, M. An overview of APP processing enzymes and products. Neuromolecular Med. 2010, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Singer, O.; Marr, R.A.; Rockenstein, E.; Crews, L.; Coufal, N.G.; Gage, F.H.; Verma, I.M.; Masliah, E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005, 8, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y. γ-Secretase in Alzheimer’s disease. Exp. Mol. Med. 2022, 54, 433–446. [Google Scholar] [CrossRef]
- Miyazaki, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Amyloid precursor protein regulates migration and metalloproteinase gene expression in prostate cancer cells. Biochem. Biophys. Res. Commun. 2014, 452, 828–833. [Google Scholar] [CrossRef]
- Zhai, K.; Huang, Z.; Huang, Q.; Tao, W.; Fang, X.; Zhang, A.; Li, X.; Stark, G.R.; Hamilton, T.A.; Bao, S. Pharmacological inhibition of BACE1 suppresses glioblastoma growth by stimulating macrophage phagocytosis of tumor cells. Nat. Cancer 2021, 2, 1136–1151. [Google Scholar] [CrossRef]
- Kleffman, K.; Levinson, G.; Rose, I.V.L.; Blumenberg, L.M.; Shadaloey, S.A.A.; Dhabaria, A.; Wong, E.; Galán-Echevarría, F.; Karz, A.; Argibay, D.; et al. Melanoma-Secreted Amyloid Beta Suppresses Neuroinflammation and Promotes Brain Metastasis. Cancer Discov. 2022, 12, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Beckett, C.; Belyaev, N.D.; Turner, A.J. The impact of amyloid precursor protein signalling and histone deacetylase inhibition on neprilysin expression in human prostate cells. Int. J. Cancer 2012, 130, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Yoo, B.K.; Kim, H.-S.; Gilmore, H.L.; Lee, Y.; Lee, H.-P.; Kim, S.-J.; Letterio, J.; Lee, H.-G. Amyloid-β precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer 2014, 14, 928. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, S.; Lu, C. Amyloid precursor protein promotes the migration and invasion of breast cancer cells by regulating the MAPK signaling pathway. Int. J. Mol. Med. 2020, 45, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Tsutsumi, S.; Suzuki, T.; Horie-Inoue, K.; Ikeda, K.; Kaneshiro, K.; Fujimura, T.; Kumagai, J.; Urano, T.; Sakaki, Y.; et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res. 2009, 69, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Zayas-Santiago, A.; Martínez-Montemayor, M.M.; Colón-Vázquez, J.; Ortiz-Soto, G.; Cirino-Simonet, J.G.; Inyushin, M. Accumulation of amyloid beta (Aβ) and amyloid precursor protein (APP) in tumors formed by a mouse xenograft model of inflammatory breast cancer. FEBS Open Bio 2022, 12, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Munir, H.; Jones, J.O.; Janowitz, T.; Hoffmann, M.; Euler, M.; Martins, C.P.; Welsh, S.J.; Shields, J.D. Stromal-driven and Amyloid β-dependent induction of neutrophil extracellular traps modulates tumor growth. Nat. Commun. 2021, 12, 683. [Google Scholar] [CrossRef]
- Farris, F.; Matafora, V.; Bachi, A. The emerging role of β-secretases in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 147. [Google Scholar] [CrossRef]
- Peters, H.L.; Tuli, A.; Wang, X.; Liu, C.; Pan, Z.; Ouellette, M.M.; Hollingsworth, M.A.; Macdonald, R.G.; Solheim, J.C. Relevance of amyloid precursor-like protein 2 C-terminal fragments in pancreatic cancer cells. Int. J. Oncol. 2012, 41, 1464–1474. [Google Scholar] [CrossRef]
- Peak, T.C.; Panigrahi, G.K.; Praharaj, P.P.; Su, Y.; Shi, L.; Chyr, J.; Rivera-Chávez, J.; Flores-Bocanegra, L.; Singh, R.; Vander Griend, D.J.; et al. Syntaxin 6-mediated exosome secretion regulates enzalutamide resistance in prostate cancer. Mol. Carcinog. 2020, 59, 62–72. [Google Scholar] [CrossRef]
- Rios-Colon, L.; Kumar, P.; Kim, S.; Sharma, M.; Su, Y.; Kumar, A.; Singh, S.; Stocks, N.; Liu, L.; Joshi, M.; et al. Carnitine Palmitoyltransferase 1 Regulates Prostate Cancer Growth under Hypoxia. Cancers 2021, 13, 6302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, P.; Sharma, M.; Kim, S.; Singh, S.; Kridel, S.J.; Deep, G. Role of extracellular vesicles secretion in paclitaxel resistance of prostate cancer cells. Cancer Drug Resist. 2022, 5, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Kemppainen, N.M.; Aalto, S.; Wilson, I.A.; Någren, K.; Helin, S.; Brück, A.; Oikonen, V.; Kailajärvi, M.; Scheinin, M.; Viitanen, M.; et al. Voxel-based analysis of PET amyloid ligand [11C]PIB uptake in Alzheimer disease. Neurology 2006, 67, 1575–1580. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, A.A.; Foster, B.A.; Allison, J.P.; Greenberg, N.M.; Kwon, E.D. The TRAMP mouse as a model for prostate cancer. Curr. Protoc. Immunol. 2001, 45, 20.5.1–20.5.23. [Google Scholar] [CrossRef]
- Kennedy, M.E.; Stamford, A.W.; Chen, X.; Cox, K.; Cumming, J.N.; Dockendorf, M.F.; Egan, M.; Ereshefsky, L.; Hodgson, R.A.; Hyde, L.A.; et al. The BACE1 inhibitor verubecestat (MK-8931) reduces CNS beta-amyloid in animal models and in Alzheimer’s disease patients. Sci. Transl. Med. 2016, 8, 363ra150. [Google Scholar] [CrossRef]
- Tiwari, R.; Manzar, N.; Ateeq, B. Dynamics of Cellular Plasticity in Prostate Cancer Progression. Front. Mol. Biosci. 2020, 7, 130. [Google Scholar] [CrossRef]
- Meethal, S.V.; Smith, M.A.; Bowen, R.L.; Atwood, C.S. The gonadotropin connection in Alzheimer’s disease. Endocrine 2005, 26, 317–326. [Google Scholar] [CrossRef]
- Bowen, R.L.; Verdile, G.; Liu, T.; Parlow, A.F.; Perry, G.; Smith, M.A.; Martins, R.N.; Atwood, C.S. Luteinizing hormone, a reproductive regulator that modulates the processing of amyloid-beta precursor protein and amyloid-beta deposition. J. Biol. Chem. 2004, 279, 20539–20545. [Google Scholar] [CrossRef]
- Kucheryavykh, L.Y.; Ortiz-Rivera, J.; Kucheryavykh, Y.V.; Zayas-Santiago, A.; Diaz-Garcia, A.; Inyushin, M.Y. Accumulation of Innate Amyloid Beta Peptide in Glioblastoma Tumors. Int. J. Mol. Sci. 2019, 20, 2482. [Google Scholar] [CrossRef]
- Bhaskar, K.; Miller, M.; Chludzinski, A.; Herrup, K.; Zagorski, M.; Lamb, B.T. The PI3K-Akt-mTOR pathway regulates Aβ oligomer induced neuronal cell cycle events. Mol. Neurodegener. 2009, 4, 14. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Zhu, J.; Wu, J.; Thompson, C.B.; Jiang, X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 31189–31197. [Google Scholar] [CrossRef] [PubMed]
- Cham, J.; Venkateswaran, A.R.; Bhangoo, M. Targeting the PI3K-AKT-mTOR Pathway in Castration Resistant Prostate Cancer: A Review Article. Clin. Genitourin. Cancer 2021, 19, 563.e1–563.e7. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Banuelos, C.A.; Imamura, Y.; Leung, J.K.; Caley, D.P.; Wang, J.; Mawji, N.R.; Sadar, M.D. Cotargeting Androgen Receptor Splice Variants and mTOR Signaling Pathway for the Treatment of Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2016, 22, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pehar, M.; Liu, Y.; Bhattacharyya, A.; Zhang, S.C.; O’Riordan, K.J.; Burger, C.; D’Adamio, L.; Puglielli, L. The amyloid precursor protein (APP) intracellular domain regulates translation of p44, a short isoform of p53, through an IRES-dependent mechanism. Neurobiol. Aging 2015, 36, 2725–2736. [Google Scholar] [CrossRef]
- Belyaev, N.D.; Kellett, K.A.; Beckett, C.; Makova, N.Z.; Revett, T.J.; Nalivaeva, N.N.; Hooper, N.M.; Turner, A.J. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem. 2010, 285, 41443–41454. [Google Scholar] [CrossRef]
- Zhang, W.; Bai, S.; Yang, J.; Zhang, Y.; Liu, Y.; Nie, J.; Meng, D.; Shi, R.; Yao, Z.; Wang, M.; et al. FoxO1 overexpression reduces Aβ production and tau phosphorylation in vitro. Neurosci. Lett. 2020, 738, 135322. [Google Scholar] [CrossRef]
- Zhang, W.; Bai, S.S.; Zhang, Q.; Shi, R.L.; Wang, H.C.; Liu, Y.C.; Ni, T.J.; Wu, Y.; Yao, Z.Y.; Sun, Y.; et al. Physalin B reduces Aβ secretion through down-regulation of BACE1 expression by activating FoxO1 and inhibiting STAT3 phosphorylation. Chin. J. Nat. Med. 2021, 19, 732–740. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rather, H.A.; Almousa, S.; Kumar, A.; Sharma, M.; Pennington, I.; Kim, S.; Su, Y.; He, Y.; Ghara, A.R.; Sai, K.K.S.; et al. The β-Secretase 1 Enzyme as a Novel Therapeutic Target for Prostate Cancer. Cancers 2024, 16, 10. https://doi.org/10.3390/cancers16010010
Rather HA, Almousa S, Kumar A, Sharma M, Pennington I, Kim S, Su Y, He Y, Ghara AR, Sai KKS, et al. The β-Secretase 1 Enzyme as a Novel Therapeutic Target for Prostate Cancer. Cancers. 2024; 16(1):10. https://doi.org/10.3390/cancers16010010
Chicago/Turabian StyleRather, Hilal A., Sameh Almousa, Ashish Kumar, Mitu Sharma, Isabel Pennington, Susy Kim, Yixin Su, Yangen He, Abdollah R. Ghara, Kiran Kumar Solingapuram Sai, and et al. 2024. "The β-Secretase 1 Enzyme as a Novel Therapeutic Target for Prostate Cancer" Cancers 16, no. 1: 10. https://doi.org/10.3390/cancers16010010




