Non-Contrast-Enhanced Multiparametric MRI of the Hypoxic Tumor Microenvironment Allows Molecular Subtyping of Breast Cancer: A Pilot Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Breast-Cancer Xenograft Model
2.2. Experimental Setup
2.3. mpMRI Protocol
2.4. Tumor Resection and Fluorescent mpIHC
2.5. Imaging Post-Processing
2.5.1. BOLD MRI
2.5.2. IVIM MRI
2.5.3. Fluorescent mpIHC
2.6. Statistical Analysis
3. Results
3.1. IVIM-MRI and BOLD-MRI Parameter Maps
3.2. mpMRI
3.2.1. BOLD-MRI
3.2.2. IVIM MRI
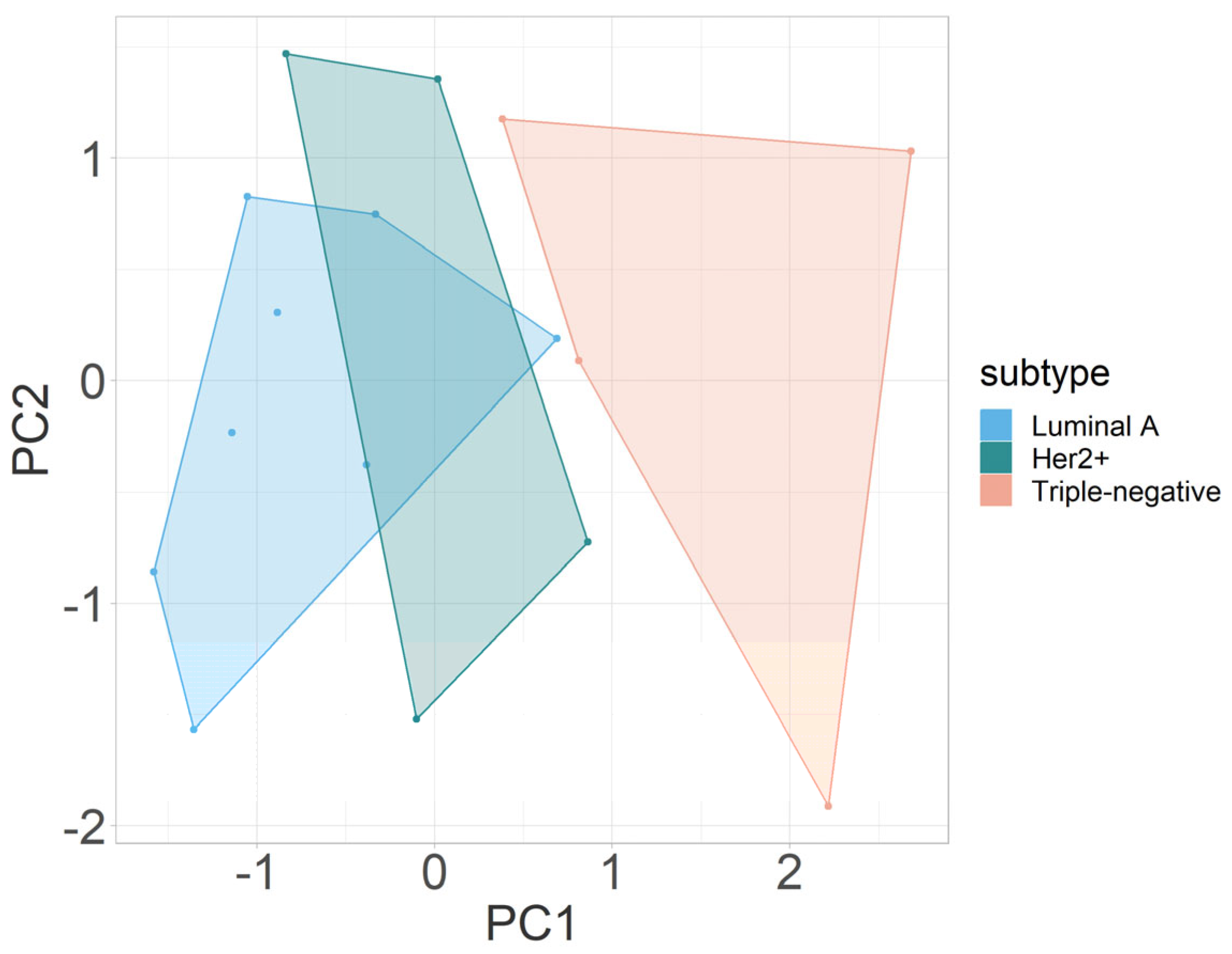
3.2.3. Multivariate Analysis
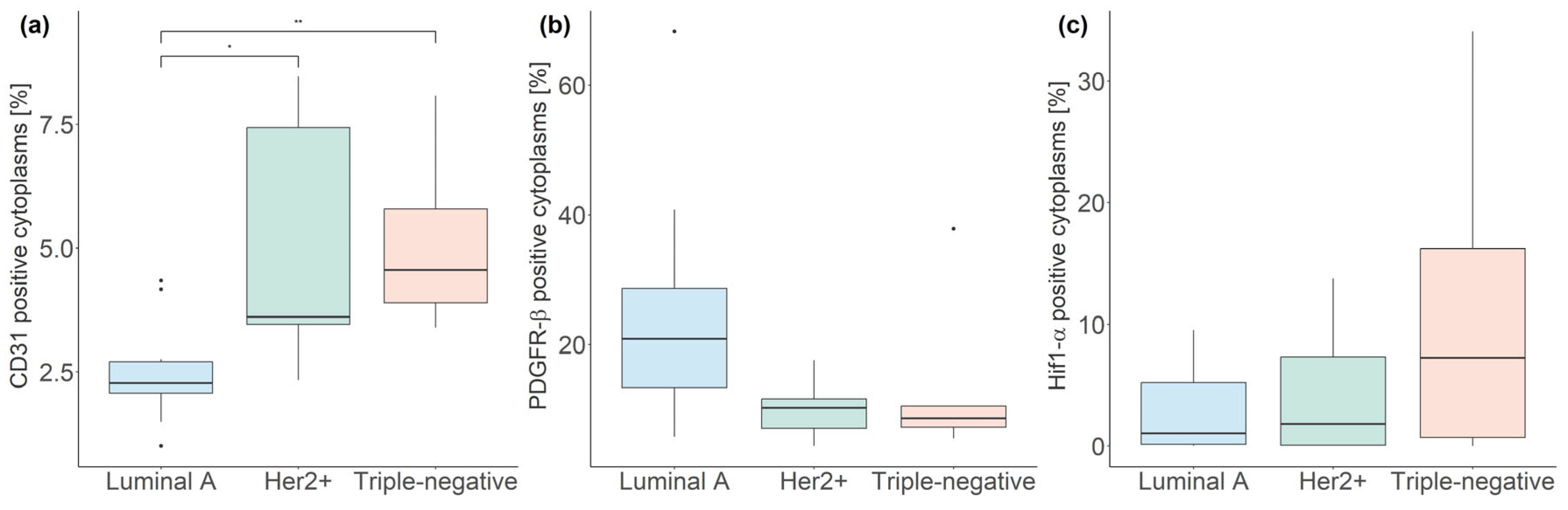
3.3. Fluorescent mpIHC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haynes, B.; Sarma, A.; Nangia-Makker, P.; Shekhar, M.P. Breast cancer complexity: Implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev. 2017, 36, 547–555. [Google Scholar] [CrossRef]
- Lüönd, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Tatum, J.L.; Kelloff, G.J.; Gillies, R.J.; Arbeit, J.M.; Brown, J.M.; Chao, K.S.; Chapman, J.D.; Eckelman, W.C.; Fyles, A.W.; Giaccia, A.J.; et al. Hypoxia: Importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int. J. Radiat. Biol. 2006, 82, 699–757. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 and tumor progression: Pathophysiology and therapeutics. Trends Mol. Med. 2001, 8, S62–S67. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Song, G.; Ouyang, G. Role of hypoxia in the hallmarks of human cancer. J. Cell. Biochem. 2009, 107, 1053–1062. [Google Scholar] [CrossRef]
- Bergers, G.; Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 2003, 3, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Uzzan, B.; Nicolas, P.; Cucherat, M.; Perret, G.-Y. Microvessel Density as a Prognostic Factor in Women with Breast Cancer: A Systematic Review of the Literature and Meta-Analysis. Cancer Res. 2004, 64, 2941–2955. [Google Scholar] [CrossRef]
- Tsutsui, S.; Kume, M.; Era, S. Prognostic value of microvessel density in invasive ductal carcinoma of the breast. Breast Cancer 2003, 10, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Denko, N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef]
- Ajduković, J. HIF-1—A big chapter in the cancer tale. Exp. Oncol. 2016, 38, 9–12. [Google Scholar] [CrossRef]
- de Heer, E.C.; Jalving, M.; Harris, A.L. HIFs, angiogenesis, and metabolism: Elusive enemies in breast cancer. J. Clin. Investig. 2020, 130, 5074–5087. [Google Scholar] [CrossRef]
- Jarman, E.J.; Ward, C.; Turnbull, A.K.; Martinez-Perez, C.; Meehan, J.; Xintaropoulou, C.; Sims, A.H.; Langdon, S.P. HER2 regulates HIF-2alpha and drives an increased hypoxic response in breast cancer. Breast Cancer Res. 2019, 21, 10. [Google Scholar] [CrossRef]
- Yang, J.; AlTahan, A.; Jones, D.T.; Buffa, F.M.; Bridges, E.; Interiano, R.B.; Qu, C.; Vogt, N.; Li, J.L.; Baban, D.; et al. Estrogen receptor-alpha directly regulates the hypoxia-inducible factor 1 pathway associated with antiestrogen response in breast cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15172–15177. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Nico, B.; Ruggieri, S.; Tamma, R.; Simone, G.; Mangia, A. Angiogenesis and Antiangiogenesis in Triple-Negative Breast cancer. Transl Oncol 2016, 9, 453–457. [Google Scholar] [CrossRef]
- Vaupel, P. Hypoxia and aggressive tumor phenotype: Implications for therapy and prognosis. Oncologist 2008, 13 (Suppl. S3), 21–26. [Google Scholar] [CrossRef]
- Pinker, K.; Bogner, W.; Baltzer, P.; Karanikas, G.; Magometschnigg, H.; Brader, P.; Gruber, S.; Bickel, H.; Dubsky, P.; Bago-Horvath, Z.; et al. Improved differentiation of benign and malignant breast tumors with multiparametric 18fluorodeoxyglucose positron emission tomography magnetic resonance imaging: A feasibility study. Clin. Cancer Res. 2014, 20, 3540–3549. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Helbich, T.H.; Morris, E.A. The potential of multiparametric MRI of the breast. Br. J. Radiol. 2017, 90, 20160715. [Google Scholar] [CrossRef]
- Bennani-Baiti, B.; Pinker, K.; Zimmermann, M.; Helbich, T.H.; Baltzer, P.A.; Clauser, P.; Kapetas, P.; Bago-Horvath, Z.; Stadlbauer, A. Non-Invasive Assessment of Hypoxia and Neovascularization with MRI for Identification of Aggressive Breast Cancer. Cancers 2020, 12, 2024. [Google Scholar] [CrossRef]
- Pinker, K.; Bickel, H.; Helbich, T.; Gruber, S.; Dubsky, P.; Pluschnig, U.; Rudas, M.; Bago-Horvath, Z.; Weber, M.; Trattnig, S. Combined contrast-enhanced magnetic resonance and diffusion-weighted imaging reading adapted to the “Breast Imaging Reporting and Data System” for multiparametric 3-T imaging of breast lesions. Eur. Radiol. 2013, 23, 1791–1802. [Google Scholar] [CrossRef]
- Rahbar, H.; Partridge, S.C. Multiparametric Breast MRI of Breast Cancer. Magn. Reson. Imaging Clin. N. Am. 2016, 24, 223–238. [Google Scholar] [CrossRef]
- Liu, M.; Guo, X.; Wang, S.; Jin, M.; Wang, Y.; Li, J.; Liu, J. BOLD-MRI of breast invasive ductal carcinoma: Correlation of R2* value and the expression of HIF-1alpha. Eur. Radiol. 2013, 23, 3221–3227. [Google Scholar] [CrossRef] [PubMed]
- Rakow-Penner, R.; Daniel, B.; Glover, G.H. Detecting blood oxygen level-dependent (BOLD) contrast in the breast. J. Magn. Reson. Imaging 2010, 32, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.J.; Ehret, V.; Friske, J.; Frohlich, V.; Laimer-Gruber, D.; Helbich, T.H.; Pinker, K. Hyperoxic BOLD-MRI-Based Characterization of Breast Cancer Molecular Subtypes Is Independent of the Supplied Amount of Oxygen: A Preclinical Study. Diagnostics 2023, 13, 2946. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Robinson, S.P.; Waterton, J.C. Imaging tumor hypoxia with oxygen-enhanced MRI and BOLD MRI. Br. J. Radiol. 2019, 92, 20180642. [Google Scholar] [CrossRef]
- Ogawa, S.; Lee, T.M.; Kay, A.R.; Tank, D.W. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef]
- Raichle, M.E. Behind the scenes of functional brain imaging: A historical and physiological perspective. Proc. Natl. Acad. Sci. USA 1998, 95, 765–772. [Google Scholar] [CrossRef]
- Le Bihan, D. What can we see with IVIM MRI? NeuroImage 2019, 187, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Le Bihan, D.; Breton, E.; Lallemand, D.; Aubin, M.-L.; Vignaud, J.; Laval-Jeantet, M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988, 168, 497–505. [Google Scholar] [CrossRef]
- Fusco, R.; Granata, V.; Mattace Raso, M.; Vallone, P.; De Rosa, A.P.; Siani, C.; Di Bonito, M.; Petrillo, A.; Sansone, M. Blood Oxygenation Level Dependent Magnetic Resonance Imaging (MRI), Dynamic Contrast Enhanced MRI, and Diffusion Weighted MRI for Benign and Malignant Breast Cancer Discrimination: A Preliminary Experience. Cancers 2021, 13, 2421. [Google Scholar] [CrossRef]
- Fusco, R.; Granata, V.; Pariante, P.; Cerciello, V.; Siani, C.; Di Bonito, M.; Valentino, M.; Sansone, M.; Botti, G.; Petrillo, A. Blood oxygenation level dependent magnetic resonance imaging and diffusion weighted MRI imaging for benign and malignant breast cancer discrimination. Magn. Reson. Imaging 2021, 75, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Iima, M. Perfusion-driven Intravoxel Incoherent Motion (IVIM) MRI in Oncology: Applications, Challenges, and Future Trends. Magn. Reson. Med. Sci. 2021, 20, 125–138. [Google Scholar] [CrossRef]
- Iima, M.; Honda, M.; Sigmund, E.E.; Ohno Kishimoto, A.; Kataoka, M.; Togashi, K. Diffusion MRI of the breast: Current status and future directions. J. Magn. Reson. Imaging 2020, 52, 70–90. [Google Scholar] [CrossRef] [PubMed]
- Harms, P.W.; Frankel, T.L.; Moutafi, M.; Rao, A.; Rimm, D.L.; Taube, J.M.; Thomas, D.; Chan, M.P.; Pantanowitz, L. Multiplex immunohistochemistry and immunofluorescence: A practical update for pathologists. Mod. Pathol. 2023, 36, 100197. [Google Scholar] [CrossRef]
- Toi, M.; Tominaga, T.; Kashitani, J. Tumor angiogenesis is an independent prognostic indicator in primary breast carcinoma. Int. J. Cancer 1993, 55, 371–374. [Google Scholar] [CrossRef]
- Zaha, D.C. Significance of immunohistochemistry in breast cancer. World J. Clin. Oncol. 2014, 5, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef]
- Hellström, M.; Gerhardt, H.; Kalén, M.; Li, X.; Eriksson, U.; Wolburg, H.; Betsholtz, C. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J. Cell Biol. 2001, 153, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Song, S. The role of pericytes in blood-vessel formation and maintenance. Neuro-Oncology 2005, 7, 452–464. [Google Scholar] [CrossRef]
- Carvalho, I.; Milanezi, F.; Martins, A.; Reis, R.M.; Schmitt, F. Overexpression of platelet-derived growth factor receptor alpha in breast cancer is associated with tumour progression. Breast Cancer Res. 2005, 7, R788–R795. [Google Scholar] [CrossRef]
- Gehmert, S.; Gehmert, S.; Prantl, L.; Vykoukal, J.; Alt, E.; Song, Y.H. Breast cancer cells attract the migration of adipose tissue-derived stem cells via the PDGF-BB/PDGFR-beta signaling pathway. Biochem. Biophys. Res. Commun. 2010, 398, 601–605. [Google Scholar] [CrossRef]
- Chiavarina, B.; Whitaker-Menezes, D.; Migneco, G.; Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Tanowitz, H.B.; Casimiro, M.C.; Wang, C.; Pestell, R.G.; et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle 2010, 9, 3534–3551. [Google Scholar] [CrossRef]
- Vleugel, M.M.; Greijer, A.E.; Shvarts, A.; van der Groep, P.; van Berkel, M.; Aarbodem, Y.; van Tinteren, H.; Harris, A.L.; van Diest, P.J.; van der Wall, E. Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J. Clin. Pathol. 2005, 58, 172–177. [Google Scholar] [CrossRef]
- Fraum, T.J.; Ludwig, D.R.; Bashir, M.R.; Fowler, K.J. Gadolinium-based contrast agents: A comprehensive risk assessment. J. Magn. Reson. Imaging 2017, 46, 338–353. [Google Scholar] [CrossRef]
- Weinreb, J.C.; Rodby, R.A.; Yee, J.; Wang, C.L.; Fine, D.; McDonald, R.J.; Perazella, M.A.; Dillman, J.R.; Davenport, M.S. Use of Intravenous Gadolinium-based Contrast Media in Patients with Kidney Disease: Consensus Statements from the American College of Radiology and the National Kidney Foundation. Radiology 2021, 298, 28–35. [Google Scholar] [CrossRef]
- Van der Molen, A.J.; Quattrocchi, C.C.; Mallio, C.A.; Dekkers, I.A.; European Society of Magnetic Resonance in Medicine, Biology Gadolinium Research, Educational Committee (ESMRMB-GREC). Ten years of gadolinium retention and deposition: ESMRMB-GREC looks backward and forward. Eur Radiol. 2023, 1–12, Erratum in Eur. Radiol. 2023. [Google Scholar] [CrossRef]
- Baltzer, A.; Dietzel, M.; Kaiser, C.G.; Baltzer, P.A. Combined reading of contrast enhanced and diffusion weighted magnetic resonance imaging by using a simple sum score. Eur. Radiol. 2016, 26, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, P.; Mann, R.M.; Iima, M.; Sigmund, E.E.; Clauser, P.; Gilbert, F.J.; Martincich, L.; Partridge, S.C.; Patterson, A.; Pinker, K. Diffusion-weighted imaging of the breast—A consensus and mission statement from the EUSOBI International Breast Diffusion-Weighted Imaging working group. Eur. Radiol. 2020, 30, 1436–1450. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Iima, M.; Kataoka, M.; Fukushima, Y.; Ota, R.; Ohashi, A.; Toi, M.; Nakamoto, Y. Biomarkers predictive of distant disease-free survival derived from diffusion-weighted imaging of breast cancer. Magn. Reson. Med. Sci. 2022, 22, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Iima, M.; Kataoka, M.; Kanao, S.; Onishi, N.; Kawai, M.; Ohashi, A.; Sakaguchi, R.; Toi, M.; Togashi, K. Intravoxel incoherent motion and quantitative non-Gaussian diffusion MR imaging: Evaluation of the diagnostic and prognostic value of several markers of malignant and benign breast lesions. Radiology 2018, 287, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Iima, M.; Nobashi, T.; Imai, H.; Koyasu, S.; Saga, T.; Nakamoto, Y.; Kataoka, M.; Yamamoto, A.; Matsuda, T.; Togashi, K. Effects of diffusion time on non-Gaussian diffusion and intravoxel incoherent motion (IVIM) MRI parameters in breast cancer and hepatocellular carcinoma xenograft models. Acta Radiol Open 2018, 7, 2058460117751565. [Google Scholar] [CrossRef]



| Subtype | IVIM MRI | BOLD MRI | ||
|---|---|---|---|---|
| D (10−3 mm2/s) | fIVIM (%) | D* (mm2/s) | ΔR2* | |
| Luminal A | 0.6350, 0.034 | 35.3, 8.6 | 0.0487, 0.026 | −28.42, 24.35 |
| Her2+ | 0.5985, 0.00673 | 33.8, 9.1 a | 0.0343, 0.042 | −2.85, 21.95 † |
| Triple-negative | 0.6240, 0.0063 | 19.5, 11.9 | 0.0468, 0.011 | −11.64, 9.15 † |
| Parameter | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| fIVIM | 0.6716729 | −0.3464966 | −0.2224006 | 0.6159007 |
| D* | −0.1683178 | −0.7922028 | −0.4168883 | −0.4126595 |
| D | −0.4323694 | −0.4692890 | 0.6342436 | 0.4365314 |
| ΔR2* | −0.5775650 | 0.1792278 | −0.6119454 | 0.5097243 |
| Subtype | CD31 (%) | PDGFR-β (%) | Hif1-α (%) |
|---|---|---|---|
| Luminal A | 2.27, 0.64 | 20.89, 15.29 | 1.04, 5.07 |
| Her2+ | 3.61, 3.97 * | 10.25, 4.51 | 1.82, 7.28 |
| Triple-negative | 4.56, 1.89 ** | 8.59, 3.24 | 7.26, 15.51 |
| Parameter | PC1 | PC2 | PC3 |
|---|---|---|---|
| PDGFR-β | 0.6791799 | 0.410240 | 0.608620 |
| Hif1-α | −0.0863886 | 0.868131 | −0.488759 |
| CD31 | −0.7288701 | 0.279378 | 0.625057 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartsch, S.J.; Brožová, K.; Ehret, V.; Friske, J.; Fürböck, C.; Kenner, L.; Laimer-Gruber, D.; Helbich, T.H.; Pinker, K. Non-Contrast-Enhanced Multiparametric MRI of the Hypoxic Tumor Microenvironment Allows Molecular Subtyping of Breast Cancer: A Pilot Study. Cancers 2024, 16, 375. https://doi.org/10.3390/cancers16020375
Bartsch SJ, Brožová K, Ehret V, Friske J, Fürböck C, Kenner L, Laimer-Gruber D, Helbich TH, Pinker K. Non-Contrast-Enhanced Multiparametric MRI of the Hypoxic Tumor Microenvironment Allows Molecular Subtyping of Breast Cancer: A Pilot Study. Cancers. 2024; 16(2):375. https://doi.org/10.3390/cancers16020375
Chicago/Turabian StyleBartsch, Silvester J., Klára Brožová, Viktoria Ehret, Joachim Friske, Christoph Fürböck, Lukas Kenner, Daniela Laimer-Gruber, Thomas H. Helbich, and Katja Pinker. 2024. "Non-Contrast-Enhanced Multiparametric MRI of the Hypoxic Tumor Microenvironment Allows Molecular Subtyping of Breast Cancer: A Pilot Study" Cancers 16, no. 2: 375. https://doi.org/10.3390/cancers16020375
APA StyleBartsch, S. J., Brožová, K., Ehret, V., Friske, J., Fürböck, C., Kenner, L., Laimer-Gruber, D., Helbich, T. H., & Pinker, K. (2024). Non-Contrast-Enhanced Multiparametric MRI of the Hypoxic Tumor Microenvironment Allows Molecular Subtyping of Breast Cancer: A Pilot Study. Cancers, 16(2), 375. https://doi.org/10.3390/cancers16020375









