Amino Terminal Acetylation of HOXB13 Regulates the DNA Damage Response in Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Chemicals
2.3. Antibodies
2.4. DNA Damage Analysis
2.5. Cell Transfection
2.6. Immunoblotting Analysis
2.7. Immunofluorescence Analysis
2.8. Clonogenic Survival Assay
2.9. Cell Proliferation Assay
2.10. DNA Fibre Assay
2.11. Flow Cytometry
2.12. Cell Cycle Analysis
2.13. Statistical Analysis
3. Results
3.1. Acetylation of HOXB13 Is Induced by DNA Damage and Persists despite PARP Inhibition
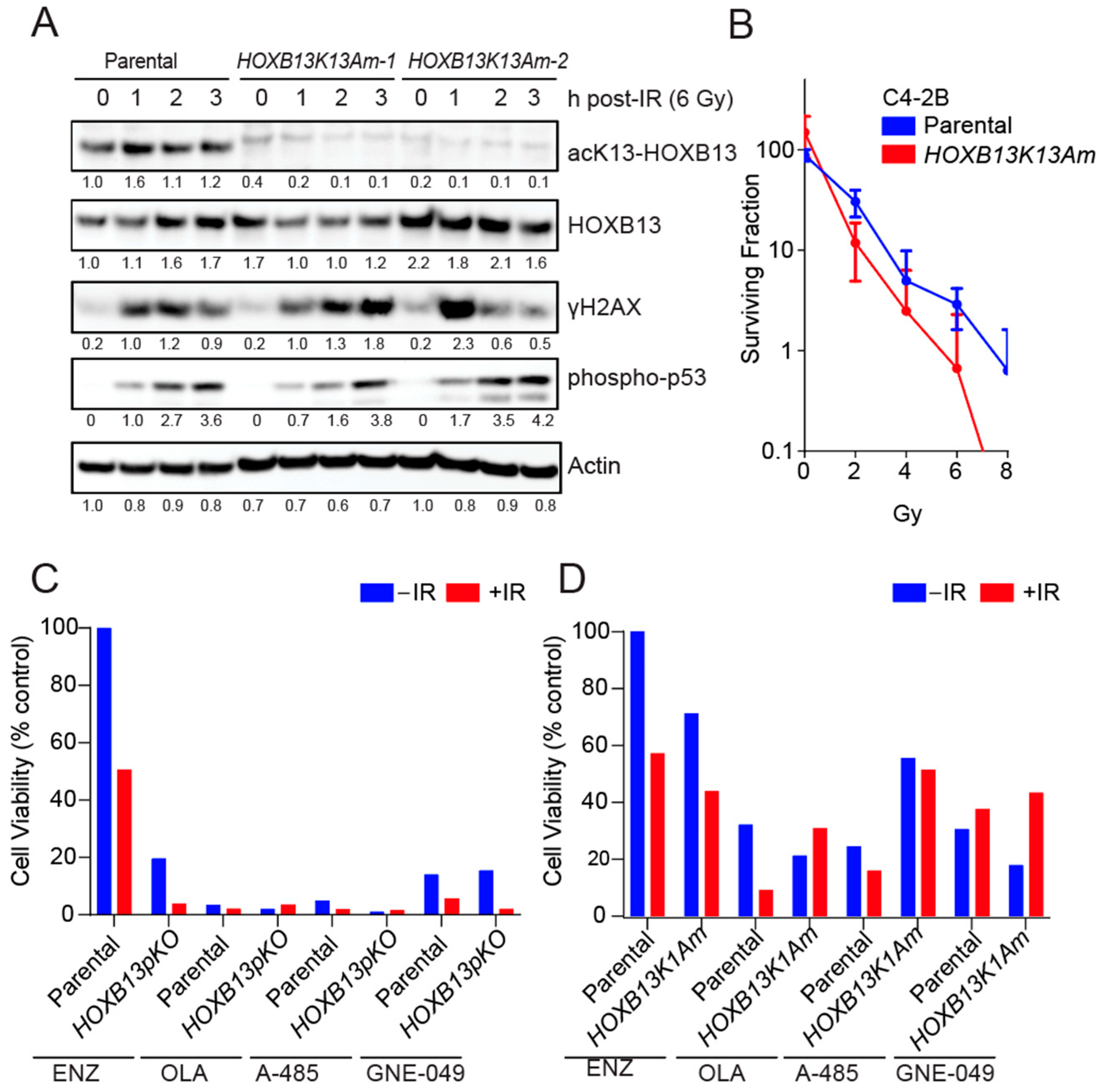
3.2. HOXB13 Acetylation Promotes Radioresistance of PC Cells
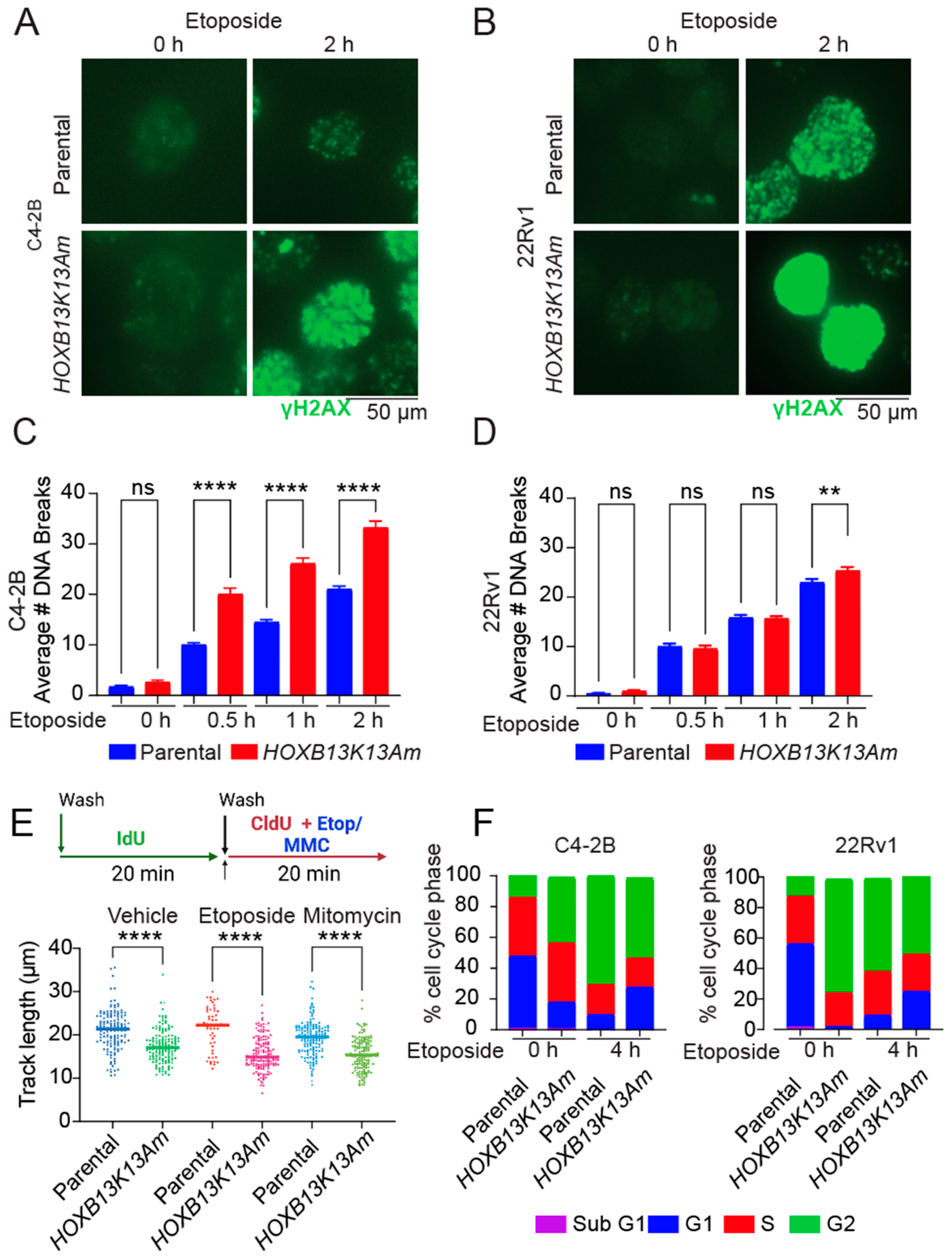
3.3. HOXB13-K13 Acetylation-Defective Mutants Show Increased DNA Damage and Impaired Replication Fork Progression
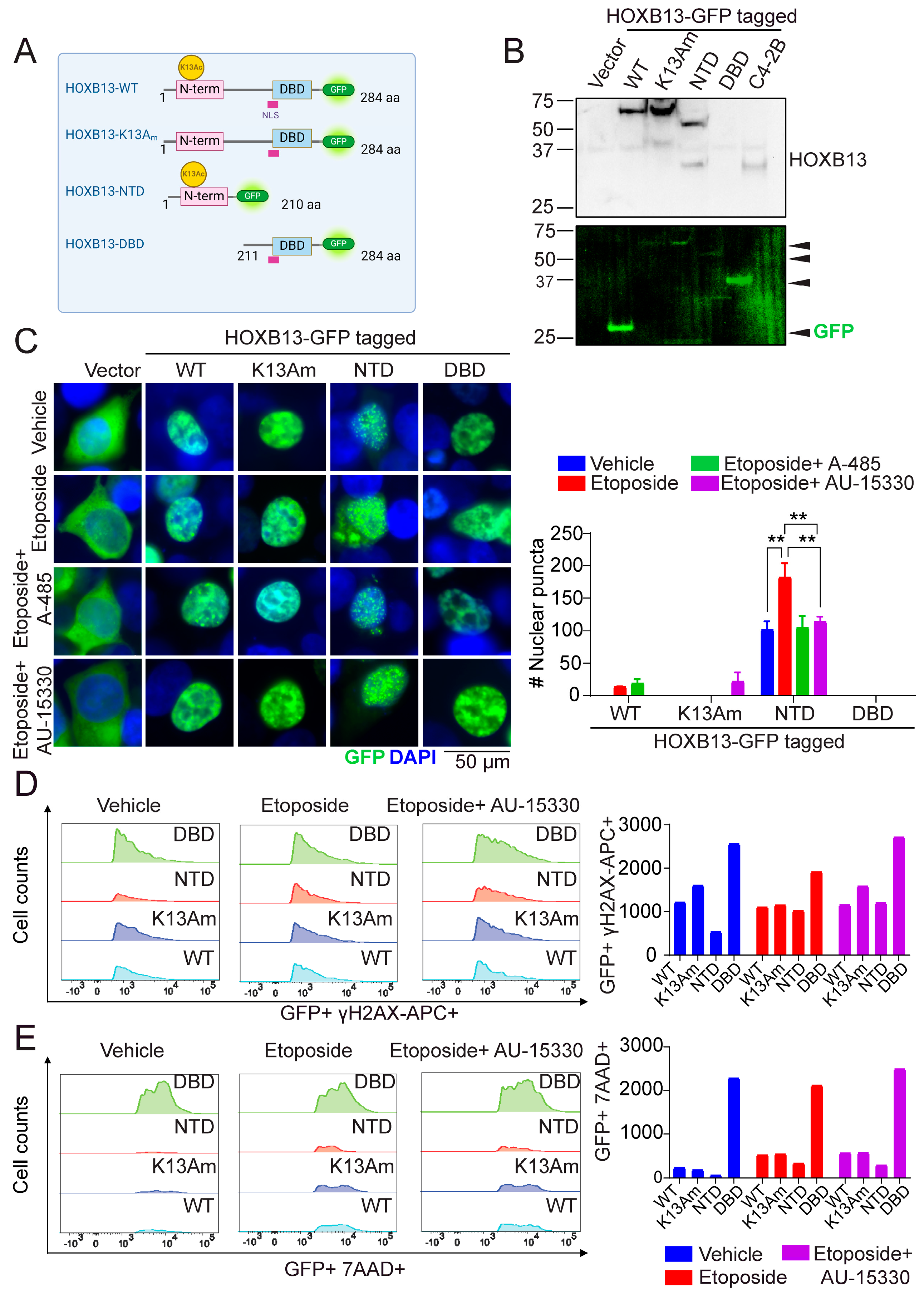
3.4. N-Terminal Disordered Region of HOXB13 Forms Nuclear Puncta and Confers Resistance to DNA Damage
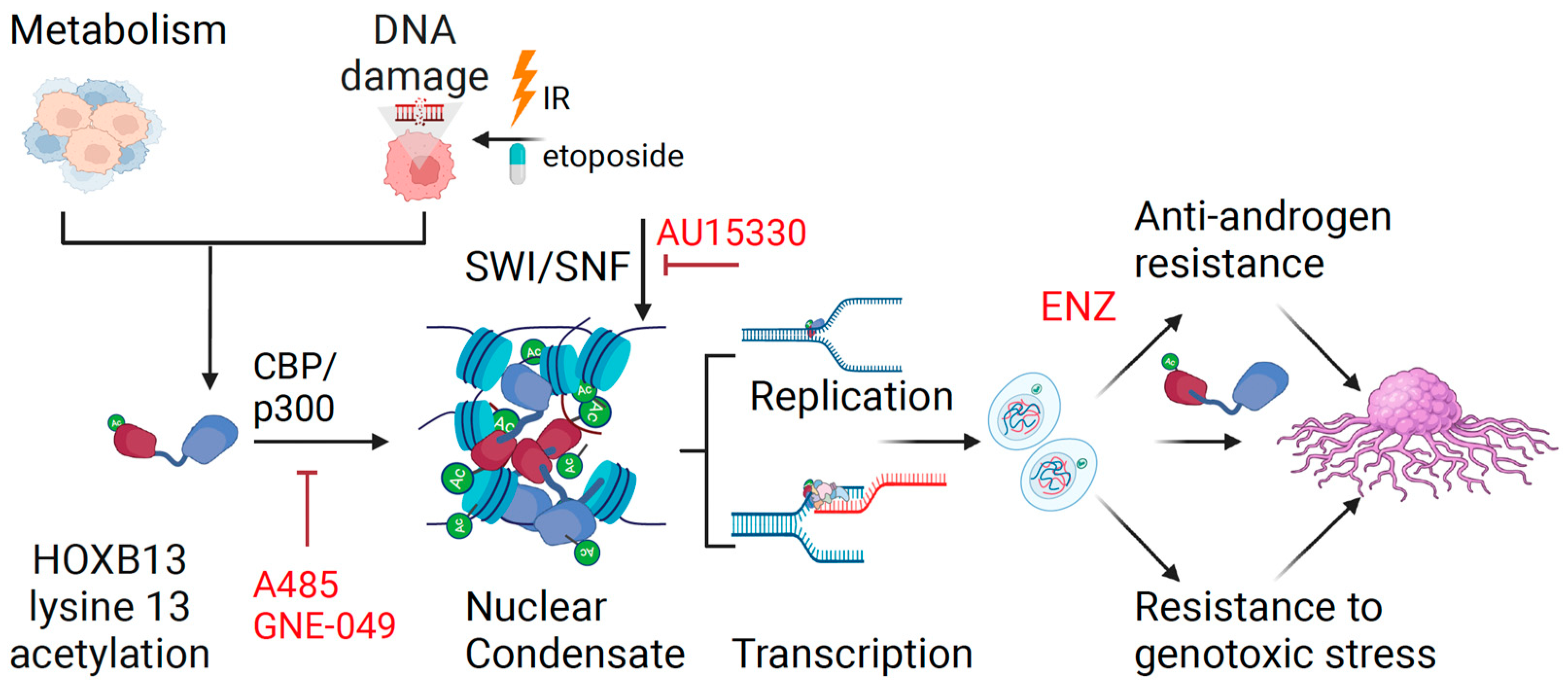
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Macedo-Silva, C.; Benedetti, R.; Ciardiello, F.; Cappabianca, S.; Jeronimo, C.; Altucci, L. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin. Epigenet. 2021, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Rusarova, N.; Vitaskova, D.; Kalabova, H.; Ondruskova, A.; Purova, D.; Melichar, B.; Studentova, H. The role of carboplatin in combination with paclitaxel in patients with castration-resistant prostate cancer. Future Oncol. 2022, 18, 4183–4192. [Google Scholar] [CrossRef]
- Cattrini, C.; Capaia, M.; Boccardo, F.; Barboro, P. Etoposide and topoisomerase II inhibition for aggressive prostate cancer: Data from a translational study. Cancer Treat. Res. Commun. 2020, 25, 100221. [Google Scholar] [CrossRef]
- Schmid, S.; Omlin, A.; Higano, C.; Sweeney, C.; Martinez Chanza, N.; Mehra, N.; Kuppen, M.C.P.; Beltran, H.; Conteduca, V.; Vargas Pivato de Almeida, D.; et al. Activity of Platinum-Based Chemotherapy in Patients with Advanced Prostate Cancer with and without DNA Repair Gene Aberrations. JAMA Netw. Open 2020, 3, e2021692. [Google Scholar] [CrossRef]
- Song, H.; Weinstein, H.N.W.; Allegakoen, P.; Wadsworth, M.H., 2nd; Xie, J.; Yang, H.; Castro, E.A.; Lu, K.L.; Stohr, B.A.; Feng, F.Y.; et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat. Commun. 2022, 13, 141. [Google Scholar] [CrossRef]
- Crowley, L.; Shen, M.M. Heterogeneity and complexity of the prostate epithelium: New findings from single-cell RNA sequencing studies. Cancer Lett. 2022, 525, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Narita, T.; Satpathy, S.; Srinivasan, B.; Hansen, B.K.; Scholz, C.; Hamilton, W.B.; Zucconi, B.E.; Wang, W.W.; Liu, W.R.; et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018, 174, 231–244 e212. [Google Scholar] [CrossRef]
- Nerlakanti, N.; Yao, J.; Nguyen, D.T.; Patel, A.K.; Eroshkin, A.M.; Lawrence, H.R.; Ayaz, M.; Kuenzi, B.M.; Agarwal, N.; Chen, Y.; et al. Targeting the BRD4-HOXB13 coregulated transcriptional networks with bromodomain-kinase inhibitors to suppress metastatic castration-resistant prostate cancer. Mol. Cancer Ther. 2018, 17, 2796–2810. [Google Scholar] [CrossRef]
- Economides, K.D.; Capecchi, M.R. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 2003, 130, 2061–2069. [Google Scholar] [CrossRef]
- Norris, J.D.; Chang, C.Y.; Wittmann, B.M.; Kunder, R.S.; Cui, H.; Fan, D.; Joseph, J.D.; McDonnell, D.P. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell 2009, 36, 405–416. [Google Scholar] [CrossRef]
- Zabalza, C.V.; Adam, M.; Burdelski, C.; Wilczak, W.; Wittmer, C.; Kraft, S.; Krech, T.; Steurer, S.; Koop, C.; Hube-Magg, C.; et al. HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy. Oncotarget 2015, 6, 12822–12834. [Google Scholar] [CrossRef]
- Weiner, A.B.; Faisal, F.A.; Davicioni, E.; Karnes, R.J.; Griend, D.J.V.; Lotan, T.L.; Schaeffer, E.M. Somatic HOXB13 Expression Correlates with Metastatic Progression in Men with Localized Prostate Cancer Following Radical Prostatectomy. Eur. Urol. Oncol. 2020, 4, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Chen, Y.; Nguyen, D.T.; Thompson, Z.J.; Eroshkin, A.M.; Nerlakanti, N.; Patel, A.K.; Agarwal, N.; Teer, J.K.; Dhillon, J.; et al. The Homeobox gene, HOXB13, Regulates a Mitotic Protein-Kinase Interaction Network in Metastatic Prostate Cancers. Sci. Rep. 2019, 9, 9715. [Google Scholar] [CrossRef]
- Barashi, N.S.; Li, T.; Angappulige, D.H.; Zhang, B.; O’Gorman, H.; Nottingham, C.U.; Shetty, A.S.; Ippolito, J.E.; Andriole, G.L.; Mahajan, N.P.; et al. Symptomatic Benign Prostatic Hyperplasia with Suppressed Epigenetic Regulator HOXB13 Shows a Lower Incidence of Prostate Cancer Development. Cancers 2024, 16, 213. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.; Yang, W.; Renganathan, A.; Weimholt, C.; Angappulige, D.H.; Nguyen, T.; Sprung, R.W.; Andriole, G.L.; Kim, E.H.; Mahajan, N.P.; et al. Acetylated HOXB13 Regulated Super Enhancer Genes Define Therapeutic Vulnerabilities of Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2022, 28, 4131–4145. [Google Scholar] [CrossRef] [PubMed]
- Angappulige, D.H.; Mahajan, N.P.; Mahajan, K. Epigenetic underpinnings of tumor-immune dynamics in prostate cancer immune suppression. Trends Cancer 2024, 10, P369–P381. [Google Scholar] [CrossRef] [PubMed]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St. Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288.e20. [Google Scholar] [CrossRef]
- Kadoch, C.; Crabtree, G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015, 1, e1500447. [Google Scholar] [CrossRef]
- Abe, Y.; Rozqie, R.; Matsumura, Y.; Kawamura, T.; Nakaki, R.; Tsurutani, Y.; Tanimura-Inagaki, K.; Shiono, A.; Magoori, K.; Nakamura, K.; et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 2015, 6, 7052. [Google Scholar] [CrossRef] [PubMed]
- Frederick, M.A.; Williamson, K.E.; Fernandez Garcia, M.; Ferretti, M.B.; McCarthy, R.L.; Donahue, G.; Luzete Monteiro, E.; Takenaka, N.; Reynaga, J.; Kadoch, C.; et al. A pioneer factor locally opens compacted chromatin to enable targeted ATP-dependent nucleosome remodeling. Nat. Struct. Mol. Biol. 2023, 30, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Baptista, T.; Grunberg, S.; Minoungou, N.; Koster, M.J.E.; Timmers, H.T.M.; Hahn, S.; Devys, D.; Tora, L. SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol. Cell 2017, 68, 130–143.e5. [Google Scholar] [CrossRef] [PubMed]
- Warfield, L.; Ramachandran, S.; Baptista, T.; Devys, D.; Tora, L.; Hahn, S. Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Mol. Cell 2017, 68, 118–129.e5. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Zou, L. Hallmarks of DNA replication stress. Mol. Cell 2022, 82, 2298–2314. [Google Scholar] [CrossRef]
- Vrtis, K.B.; Dewar, J.M.; Chistol, G.; Wu, R.A.; Graham, T.G.W.; Walter, J.C. Single-strand DNA breaks cause replisome disassembly. Mol. Cell 2021, 81, 1309–1318.e6. [Google Scholar] [CrossRef]
- Mahajan, K.; Malla, P.; Lawrence, H.R.; Chen, Z.; Kumar-Sinha, C.; Malik, R.; Shukla, S.; Kim, J.; Coppola, D.; Lawrence, N.J.; et al. ACK1/TNK2 Regulates Histone H4 Tyr88-phosphorylation and AR Gene Expression in Castration-Resistant Prostate Cancer. Cancer Cell 2017, 31, 790–803.e8. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.; Kastan, M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef]
- Quinet, A.; Carvajal-Maldonado, D.; Lemacon, D.; Vindigni, A. DNA Fiber Analysis: Mind the Gap! Methods Enzymol. 2017, 591, 55–82. [Google Scholar] [CrossRef] [PubMed]
- Quinet, A.; Lemacon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Bridoux, L.; Gofflot, F.; Rezsohazy, R. HOX Protein Activity Regulation by Cellular Localization. J. Dev. Biol. 2021, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Hankey, W.; Chen, Z.; Wang, Q. Shaping Chromatin States in Prostate Cancer by Pioneer Transcription Factors. Cancer Res. 2020, 80, 2427–2436. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.; Paterson, M.C. Defective DNA repair and increased lethality in ataxia telangiectasia cells exposed to 4-nitroquinoline-1-oxide. Nature 1980, 287, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.R.; Pacold, M.E.; Huang, Q.; Clarke, S.M.; Lam, F.C.; Cannell, I.G.; Bryson, B.D.; Rameseder, J.; Lee, M.J.; Blake, E.J.; et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 2013, 498, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Lai, C.H.; Zhao, X.; Saito, S.; Hamilton, M.H.; Appella, E.; Yao, T.P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001, 20, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.G.; Chen, W.S.; Das, R.; Chang, S.L.; Tomlins, S.A.; Chou, J.; Quigley, D.A.; Dang, H.X.; Barnard, T.J.; Mahal, B.A.; et al. Clinical and Genomic Implications of Luminal and Basal Subtypes Across Carcinomas. Clin. Cancer Res. 2019, 25, 2450–2457. [Google Scholar] [CrossRef]
- Li, X.; Baek, G.; Carreira, S.; Yuan, W.; Ma, S.; Hofstad, M.; Lee, S.; Gao, Y.; Bertan, C.; Fenor de la Maza, M.L.D.; et al. Targeting radioresistance and replication fork stability in prostate cancer. JCI Insight 2022, 7, e152955. [Google Scholar] [CrossRef]
- Sheahan, A.V.; Ellis, L. Epigenetic reprogramming: A key mechanism driving therapeutic resistance. Urol. Oncol. 2018, 36, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewski, T.; Prakash, S.; Roule, T.; Wagner, D. The Role and Activity of SWI/SNF Chromatin Remodelers. Annu. Rev. Plant Biol. 2023, 74, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Parolia, A.; Qiao, Y.; Bawa, P.; Eyunni, S.; Mannan, R.; Carson, S.E.; Chang, Y.; Wang, X.; Zhang, Y.; et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 2022, 601, 434–439. [Google Scholar] [CrossRef] [PubMed]
- St Pierre, R.; Kadoch, C. Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 2017, 42, 56–67. [Google Scholar] [CrossRef] [PubMed]
- de Miguel, F.J.; Gentile, C.; Feng, W.W.; Silva, S.J.; Sankar, A.; Exposito, F.; Cai, W.L.; Melnick, M.A.; Robles-Oteiza, C.; Hinkley, M.M.; et al. Mammalian SWI/SNF chromatin remodeling complexes promote tyrosine kinase inhibitor resistance in EGFR-mutant lung cancer. Cancer Cell 2023, 41, 1516–1534.e9. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Shirnekhi, H.K.; Gorman, S.D.; Chandra, B.; Baggett, D.W.; Park, C.G.; Somjee, R.; Lang, B.; Hosseini, S.M.H.; Pioso, B.J.; et al. Defining the condensate landscape of fusion oncoproteins. Nat. Commun. 2023, 14, 6008. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e16. [Google Scholar] [CrossRef]
- Shrinivas, K.; Sabari, B.R.; Coffey, E.L.; Klein, I.A.; Boija, A.; Zamudio, A.V.; Schuijers, J.; Hannett, N.M.; Sharp, P.A.; Young, R.A.; et al. Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 2019, 75, 549–561.e7. [Google Scholar] [CrossRef]
- Zamudio, A.V.; Dall’Agnese, A.; Henninger, J.E.; Manteiga, J.C.; Afeyan, L.K.; Hannett, N.M.; Coffey, E.L.; Li, C.H.; Oksuz, O.; Sabari, B.R.; et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol. Cell 2019, 76, 753–766.e6. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.T.; Mahajan, U.; Angappulige, D.H.; Doshi, A.; Mahajan, N.P.; Mahajan, K. Amino Terminal Acetylation of HOXB13 Regulates the DNA Damage Response in Prostate Cancer. Cancers 2024, 16, 1622. https://doi.org/10.3390/cancers16091622
Nguyen DT, Mahajan U, Angappulige DH, Doshi A, Mahajan NP, Mahajan K. Amino Terminal Acetylation of HOXB13 Regulates the DNA Damage Response in Prostate Cancer. Cancers. 2024; 16(9):1622. https://doi.org/10.3390/cancers16091622
Chicago/Turabian StyleNguyen, Duy T., Urvashi Mahajan, Duminduni Hewa Angappulige, Aashna Doshi, Nupam P. Mahajan, and Kiran Mahajan. 2024. "Amino Terminal Acetylation of HOXB13 Regulates the DNA Damage Response in Prostate Cancer" Cancers 16, no. 9: 1622. https://doi.org/10.3390/cancers16091622




