New Tailor-Made Alkyl-Aldehyde Bifunctional Supports for Lipase Immobilization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation of New Alkyl-Aldehyde Supports
2.2. Immobilization of LipC12 on New Alkyl-Aldehyde Supports
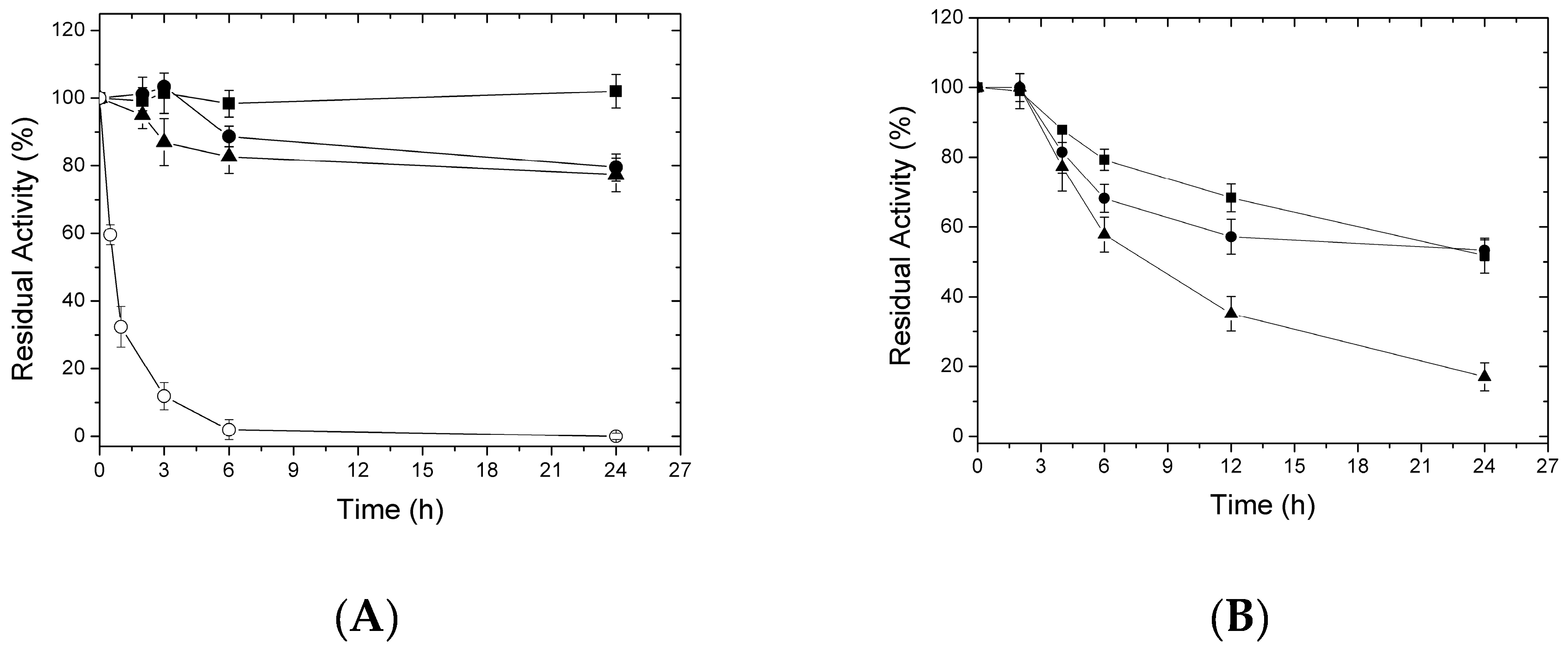
2.3. Thermal Inactivation of Different Immobilized LipC12 Preparations
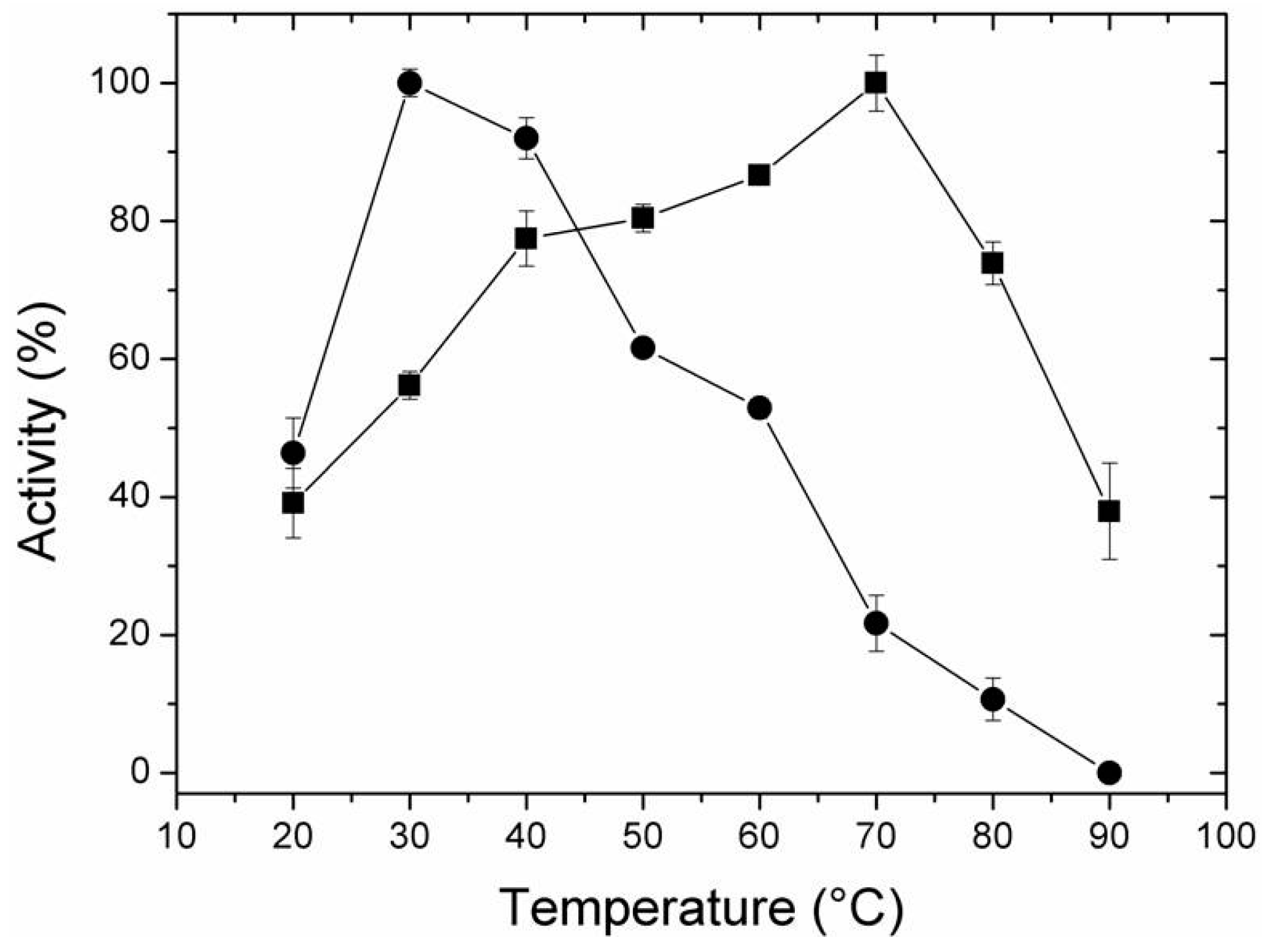
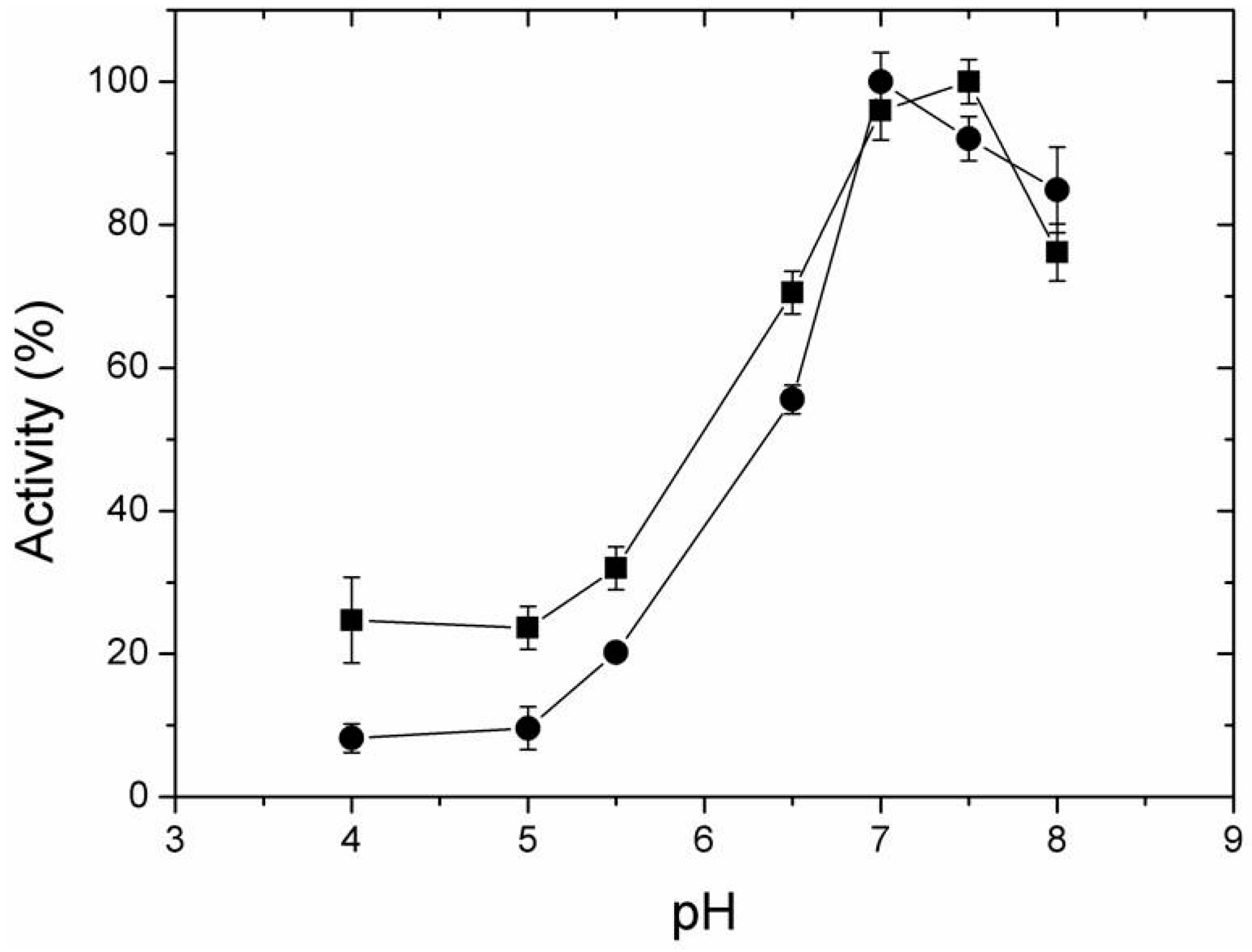
2.4. Effect of Temperature and pH on Activity of Free and Immobilized LipC12
2.5. Regioselective Hydrolysis of 3,4,6-tri-O-acetyl-d-glucal by Immobilized LipC12
3. Materials and Methods
3.1. Materials
3.2. Overexpression of Recombinant LipC12
3.3. Protein Content Determination and Electrophoresis Analysis
3.4. Lipase Activity Assay
3.5. Preparation of Supports
3.5.1. Epoxy-Agarose
3.5.2. Epoxy-Agarose-IDA-Ni2+
3.5.3. Alkyl-Agarose-Aldehyde
3.6. Purification of Recombinant LipC12
3.7. Enzyme Immobilization
3.8. Thermal Stability
3.9. Effect of pH and Temperature on the Activity of Free and Immobilized LipC12
3.10. Hydrolysis of 3,4,6-tri-O-acetyl-d-glucal
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jaeger, K.-E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; van Heuvel, M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Glogauer, A.; Martini, V.P.; Faoro, H.; Couto, G.H.; Müller-Santos, M.; Monteiro, R.A.; Mitchell, D.A.; de Souza, E.M.; Pedrosa, F.O.; Krieger, N. Identification and characterization of a new true lipase isolated through metagenomic approach. Microb. Cell Fact. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the Support Properties for Immobilization or Purification of Enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Alnoch, R.C.; Martini, V.P.; Glogauer, A.; Costa, A.C.D.S.; Piovan, L.; Muller-Santos, M.; De Souza, E.M.; Pedrosa, F.D.O.; Mitchell, D.A.; Krieger, N. Immobilization and characterization of a new regioselective and enantioselective lipase obtained from a metagenomic library. PLoS ONE 2015, 10, e0114945. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; Dos Santos, C.S.; Rodriguez, M.D.; Albuquerque, T.L.; Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Reversible immobilization of lipases on octyl-glutamic agarose beads: A mixed adsorption that reinforces enzyme immobilization. J. Mol. Catal. B Enzym. 2016, 128, 10–18. [Google Scholar] [CrossRef]
- Guisan, J.M.; Sabuquillo, P.; Fernandez-Lafuente, R.; Fernandez-Lorente, G.; Mateo, C.; Halling, P.J.; Kennedy, D.; Miyata, E.; Re, D. Preparation of new lipases derivatives with high activity-stability in anhydrous media: Adsorption on hydrophobic supports plus hydrophilization with polyethylenimine. J. Mol. Catal. B Enzym. 2001, 11, 817–824. [Google Scholar] [CrossRef]
- Palomo, J.M.; Munoz, G.; Fernandez-Lorente, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Interfacial adsorption of lipases on very hydrophobic support (octadecyl-Sepabeads): immobilization, hyperactivation and stabilization of the open form of lipases. J. Mol. Catal. B Enzym. 2002, 19, 279–286. [Google Scholar] [CrossRef]
- Carrasco-Lopez, C.; Godoy, C.; de las Rivas, B.; Fernandez-Lorente, G.; Palomo, J.M.; Guisan, J.M.; Fernandez-Lafuente, R.; Martinez-Ripoll, M.; Hermoso, J.A. Activation of bacterial thermo alkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 2009, 284, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lafuente, R.; Armisen, P.; Sabuquillo, P.; Fernández-Lorente, G.; Guisán, J.M. Immobilization of lipases by selective adsorption on hydrophobic supports. Chem. Phys. Lipids 1998, 93, 185–197. [Google Scholar] [CrossRef]
- Cao, J.; Li, Y.; Tu, N.; Lv, Y.; Chen, Q.; Dong, H. Novel enzyme/exfoliated bentonite nanohybrids as highly efficient and recyclable biocatalysts in hydrolytic reaction. J. Mol. Catal. B Enzym. 2016, 132, 41–46. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, X.; Gao, Q.; Fang, F.; Wang, L.; Huang, X. Immobilization of lipase onto functional cyclomatrix polyphosphazene microspheres. J. Mol. Catal. B Enzym. 2016, 132, 67–74. [Google Scholar] [CrossRef]
- Sato, R.; Tokuyama, H. Fabrication of enzyme-entrapped composite and macroporous gel beads by suspension gelation combined with sedimentation polymerization. Biochem. Eng. J. 2016, 113, 152–157. [Google Scholar] [CrossRef]
- Isobe, N.; Lee, D.S.; Kwon, Y.J.; Kimura, S.; Kuga, S.; Wada, M.; Kim, U.J. Immobilization of protein on cellulose hydrogel. Cellulose 2011, 18, 1251–1256. [Google Scholar] [CrossRef]
- Verri, F.; Diaz, U.; MacArio, A.; Corma, A.; Giordano, G. Optimized hybrid nanospheres immobilizing Rhizomucor miehei lipase for chiral biotransformation. Process Biochem. 2016, 51, 240–248. [Google Scholar] [CrossRef]
- Mathesh, M.; Luan, B.; Akanbi, T.O.; Weber, J.K.; Liu, J.; Barrow, C.J.; Zhou, R.; Yang, W. Opening Lids: Modulation of Lipase Immobilization by Graphene Oxides. ACS Catal. 2016, 6, 4760–4768. [Google Scholar] [CrossRef]
- Yagar, H.; Balkan, U. Entrapment of laurel lipase in chitosan hydrogel beads. Artif. Cells Nanomed. Biotechnol. 2016, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Suescun, A.; Rueda, N.; Dos Santos, J.C.S.; Castillo, J.J.; Ortiz, C.; Torres, R.; Barbosa, O.; Fernandez-Lafuente, R. Immobilization of lipases on glyoxyl-octyl supports: Improved stability and reactivation strategies. Process Biochem. 2015, 50, 1211–1217. [Google Scholar] [CrossRef]
- Peirce, S.; Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Rueda, N.; Barto Lome-Cabrero, R.; Fernandez-Lopez, L.; Russo, M.E.; Marzocchella, A.; Fernandez-Lafuente, R. Development of simple protocols to solve the problems of enzyme coimmobilization. Application to coimmobilize a lipase and a β-galactosidase. RSC Adv. 2016, 6, 61707–61715. [Google Scholar] [CrossRef]
- Marciello, M.; Filice, M.; Palomo, J.M. Different strategies to enhance the activity of lipase catalysts. Catal. Sci. Technol. 2012, 2, 1531–1543. [Google Scholar] [CrossRef]
- Palomo, J.M. Lipases enantioselectivity alteration by immobilization techniques. Curr. Bioact. Compd. 2008, 4, 126–138. [Google Scholar] [CrossRef]
- Filice, M.; Guisan, J.M.; Terreni, M.; Palomo, J.M. Regioselective monodeprotection of peracetylated carbohydrates. Nat. Protoc. 2012, 7, 1783–1796. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Bolivar, J.M.; Godoy, C.A.; Rocha-Martin, J.; Pessela, B.C.; Curiel, J.A.; Munoz, R.; Guisan, J.M.; Fernandez-Lorente, G. Improvement of enzyme properties with a two-step immobilizaton process on novel heterofunctional supports. Biomacromolecules 2010, 11, 3112–3117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henley, J.P.; Sadana, A. Deactivation theory. Biotechnol. Bioeng. 1986, 28, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Segura, R.L.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Improving the activity of lipases from thermophilic organisms at mesophilic temperatures for biotechnology applications. Biomacromolecules 2004, 5, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Jigawa, K.; Henares, T.G.; Sueyoshi, K.; Endo, T.; Hisamoto, H. Efficient immobilization of the enzyme and substrate for a single-step caspase-3 inhibitor assay using a combinable PDMS capillary sensor array. RSC Adv. 2014, 4, 7682–7687. [Google Scholar] [CrossRef]
- MacArio, A.; Giordano, G.; Frontera, P.; Crea, F.; Setti, L. Hydrolysis of alkyl ester on lipase/silicalite-1 catalyst. Catal. Lett. 2008, 122, 43–52. [Google Scholar] [CrossRef]
- Ghosh, U.; Ganessunker, D.; Sattigeri, V.J.; Carlson, K.E.; Mortensen, D.J.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A. Estrogenic diazenes: Heterocyclic non-steroidal estrogens of unusual structure with selectivity for estrogen receptor subtypes. Bioorgan. Med. Chem. 2002, 11, 629–657. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Ferna ndez-Lorente, G.; Palomo, J.M.; Mateo, C.; Munilla, R.; Ortiz, C.; Cabrera, Z.; Guisan, J.M.; Fernandez-Lafuente, R. Glutaraldehyde cross-linking of lipases adsorbed on aminated supports in the presence of detergents leads to improved performance. Biomacromolecules 2006, 7, 2610–2615. [Google Scholar] [CrossRef] [PubMed]
- Whistler, R.L.; Wolfrom, M.L.; BeMiller, J.N. Methods in Carbohydrate Chemistry; Academic Press: New York, NY, USA; London, UK, 1963; Volume 2. [Google Scholar]
- Mateo, C.; Fernandez-Lorente, G.; Pessela, B.C.C.; Vian, A.; Carrascosa, A.V.; Garcia, J.L.; Fernandez-Lafuente, R.; Guisan, J.M. Affinity chromatography of polyhistidine tagged enzymes: New dextran-coated immobilized metal ion affinity chromatography matrices for prevention of undesired multipoint adsorptions. J. Chromatogr. A 2001, 915, 97–106. [Google Scholar] [CrossRef]
- Combes, D.; Yoovidhya, T.; Girbal, E.; Willemot, R.M.; Monsan, P. Mechanism of enzyme stabilization. Ann. N. Y. Acad. Sci. 1987, 501, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Addorisio, V.; Sannino, F.; Mateo, C.; Guisan, J.M. Oxidation of phenyl compounds using strongly stable immobilized-stabilized laccase from Trametes versicolor. Process Biochem. 2013, 48, 1174–1180. [Google Scholar] [CrossRef]
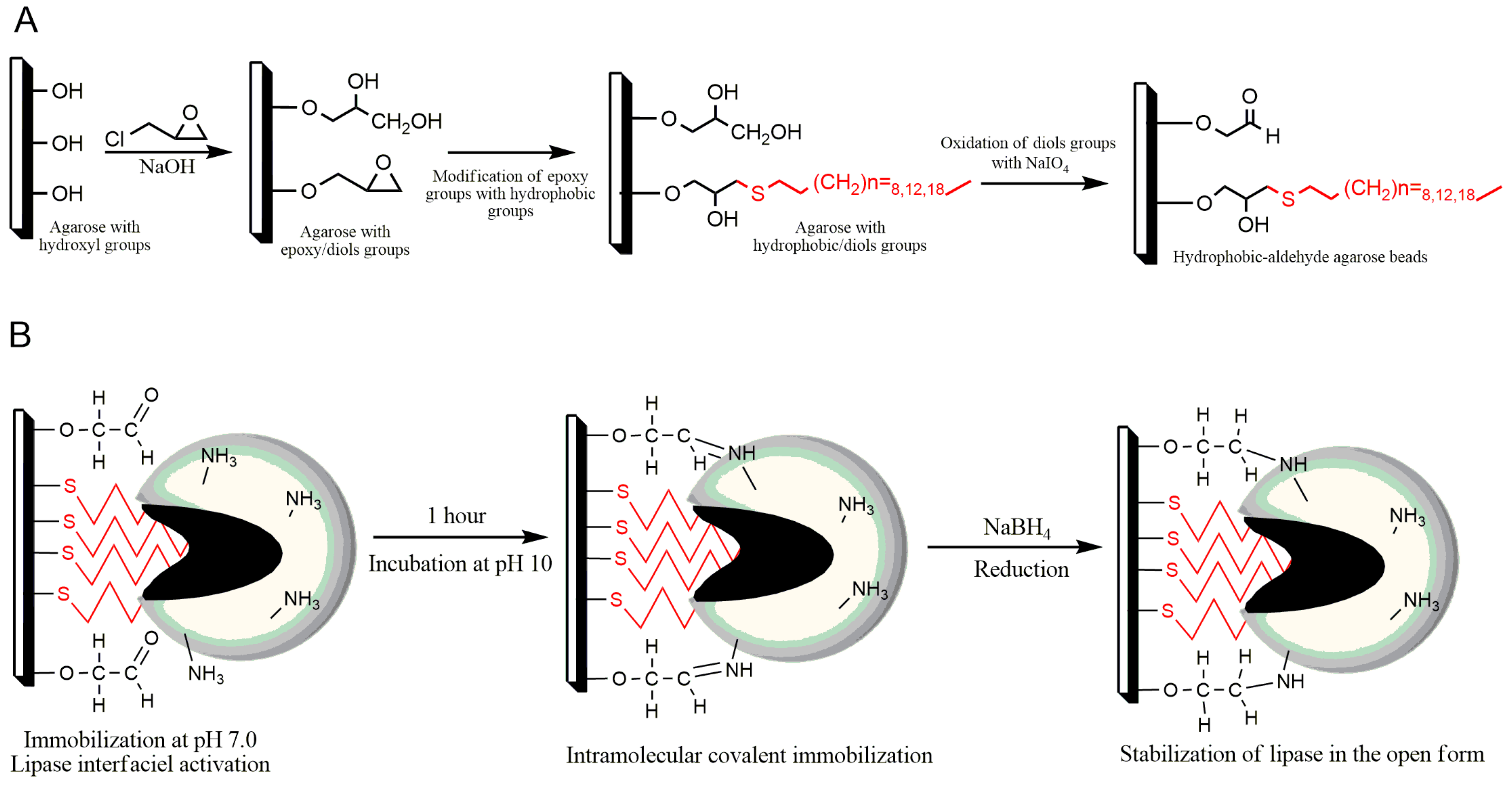

| Support | Ligands (μmol·g−1) | Diol Groups (μmol·g−1) |
|---|---|---|
| Agarose-Epoxy | 23 ± 0.4 | 43 ± 0.4 |
| C8-aldehyde | 23 ± 1 | 43 ± 1 |
| C12-aldehyde | 21 ± 1.6 | 41 ± 1.6 |
| C18-aldehyde | 19 ± 1.1 | 38 ± 1.1 |
| Support | Immobilization Efficiency (%) a | Recovered Activity b (%) | Recovered Activity after Reduction c |
|---|---|---|---|
| C8-aldehyde | >95 | 357 | 346 |
| C12-aldehyde | >95 | 380 | 370 |
| C18-aldehyde | >95 | 252 | 256 |
| Preparations a | Half-Life (t) at 80 °C | Stability Factor |
|---|---|---|
| Soluble enzyme | 0.004 | - |
| C8-aldehyde/LipC12 | 22 | 5500 |
| C12-aldehyde/LipC12 | 21 | 5250 |
| C18-aldehyde/LipC12 | 8 | 2000 |
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Preparation | pH | T °C | Specific Activity (U·mg−1) * | Time (h) | Total Conversion a (%) | Yield 2 (%) | Yield 3 (%) | Yield 4 (%) | Other Products b (%) |
| C12-aldehyde /LipC12 | 7.0 | 4 | 18 | 96 | 77 | 52 | 1 | 7 | 17 |
| C12-aldehyde /LipC12 | 5.0 | 4 | 4 | 96 | 34 | 26 | 1 | 1 | 6 |
| C12-aldehyde /LipC12-PEG | 5.0 | 4 | 15 | 96 | 81 | 69 | 0 | 3 | 9 |
| C12-aldehyde /LipC12 | 7.0 | 25 | 140 | 24 | 100 | 5 | 4 | 0 | 91 |
| C12-aldehyde /LipC12 | 5.0 | 25 | 140 | 24 | 100 | 11 | 0 | 0 | 89 |
| C12-aldehyde /LipC12-PEG | 5.0 | 25 | 110 | 24 | 100 | 22 | 2 | 2 | 74 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alnoch, R.C.; Rodrigues de Melo, R.; Palomo, J.M.; Maltempi de Souza, E.; Krieger, N.; Mateo, C. New Tailor-Made Alkyl-Aldehyde Bifunctional Supports for Lipase Immobilization. Catalysts 2016, 6, 191. https://doi.org/10.3390/catal6120191
Alnoch RC, Rodrigues de Melo R, Palomo JM, Maltempi de Souza E, Krieger N, Mateo C. New Tailor-Made Alkyl-Aldehyde Bifunctional Supports for Lipase Immobilization. Catalysts. 2016; 6(12):191. https://doi.org/10.3390/catal6120191
Chicago/Turabian StyleAlnoch, Robson Carlos, Ricardo Rodrigues de Melo, Jose M. Palomo, Emanuel Maltempi de Souza, Nadia Krieger, and Cesar Mateo. 2016. "New Tailor-Made Alkyl-Aldehyde Bifunctional Supports for Lipase Immobilization" Catalysts 6, no. 12: 191. https://doi.org/10.3390/catal6120191






