Assessing the Population Structure of Colorado Potato Beetle Populations in Croatia Using Genetic and Geometric Morphometric Tools
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Genetic Analyses
2.3. Geometric Morphometric Analyses
3. Results
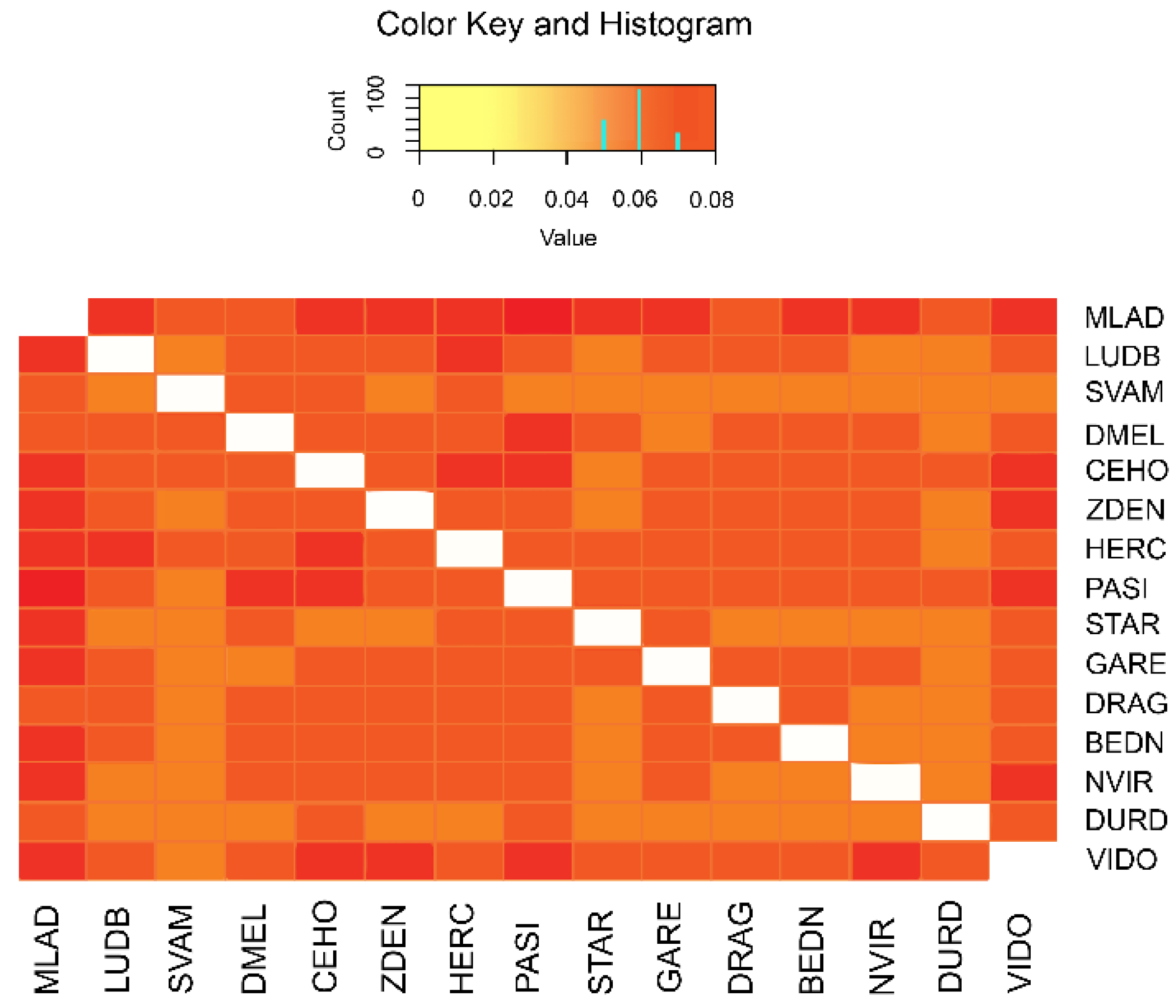
3.1. Genetic Variability
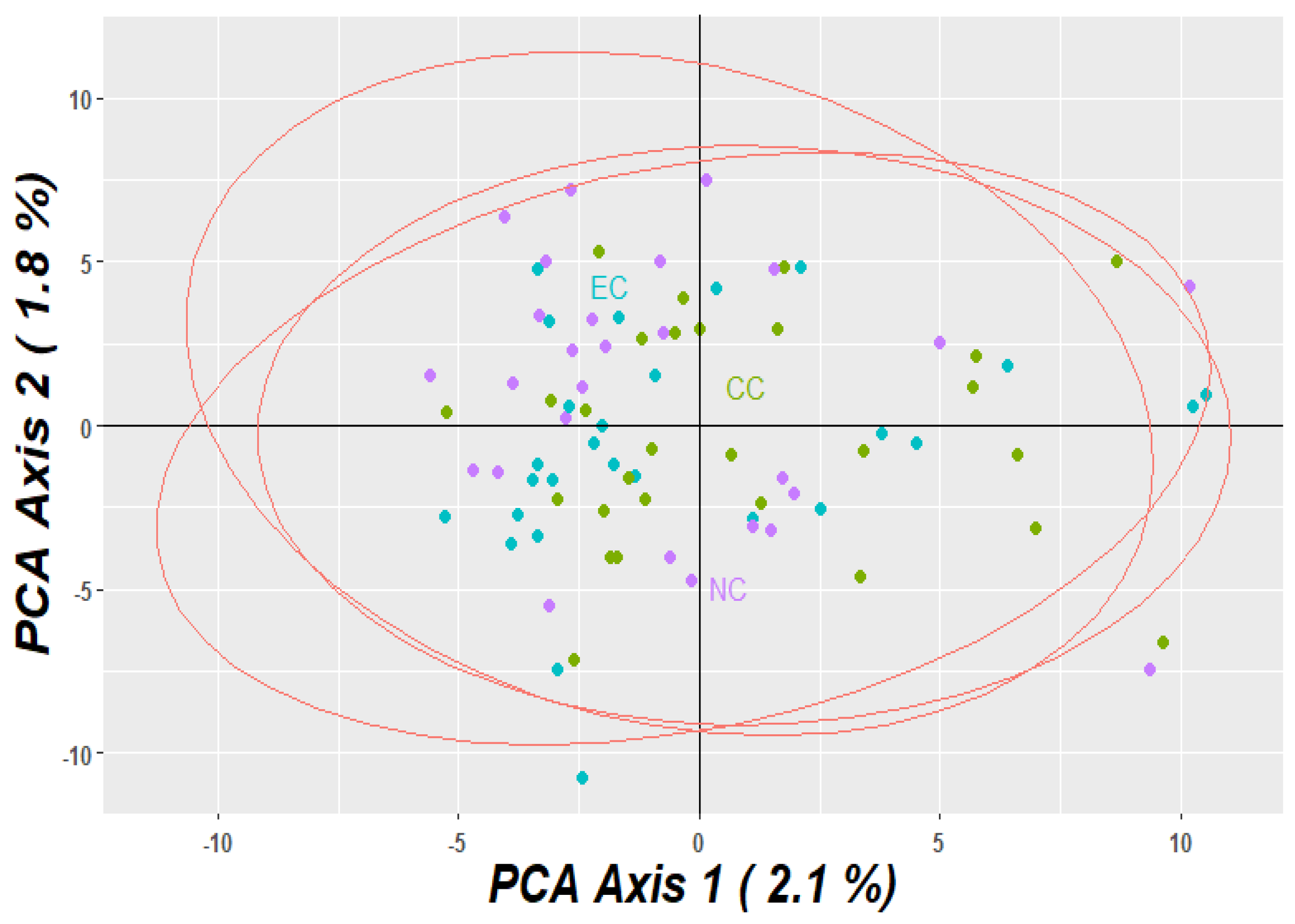
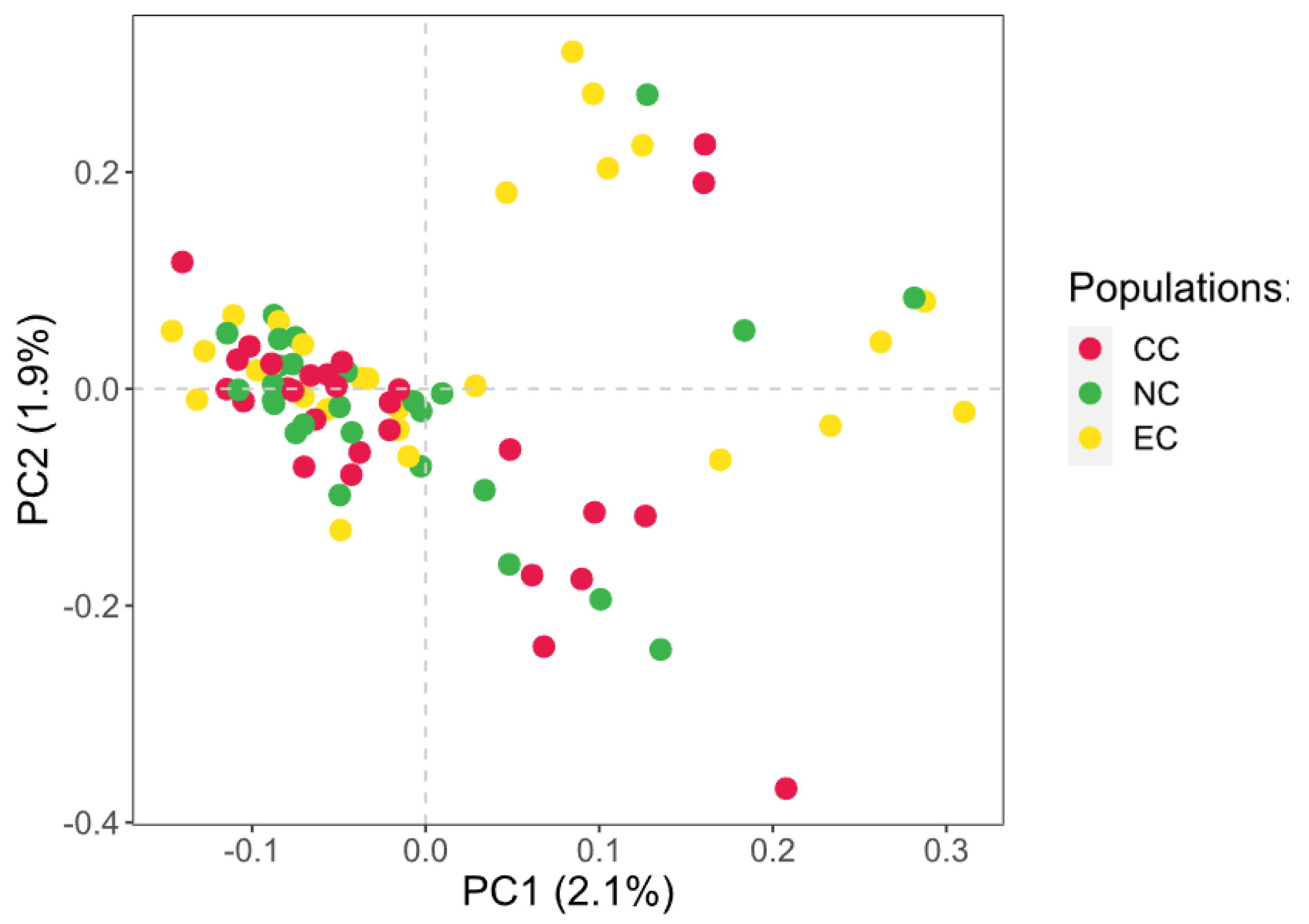
3.2. Population Relationship
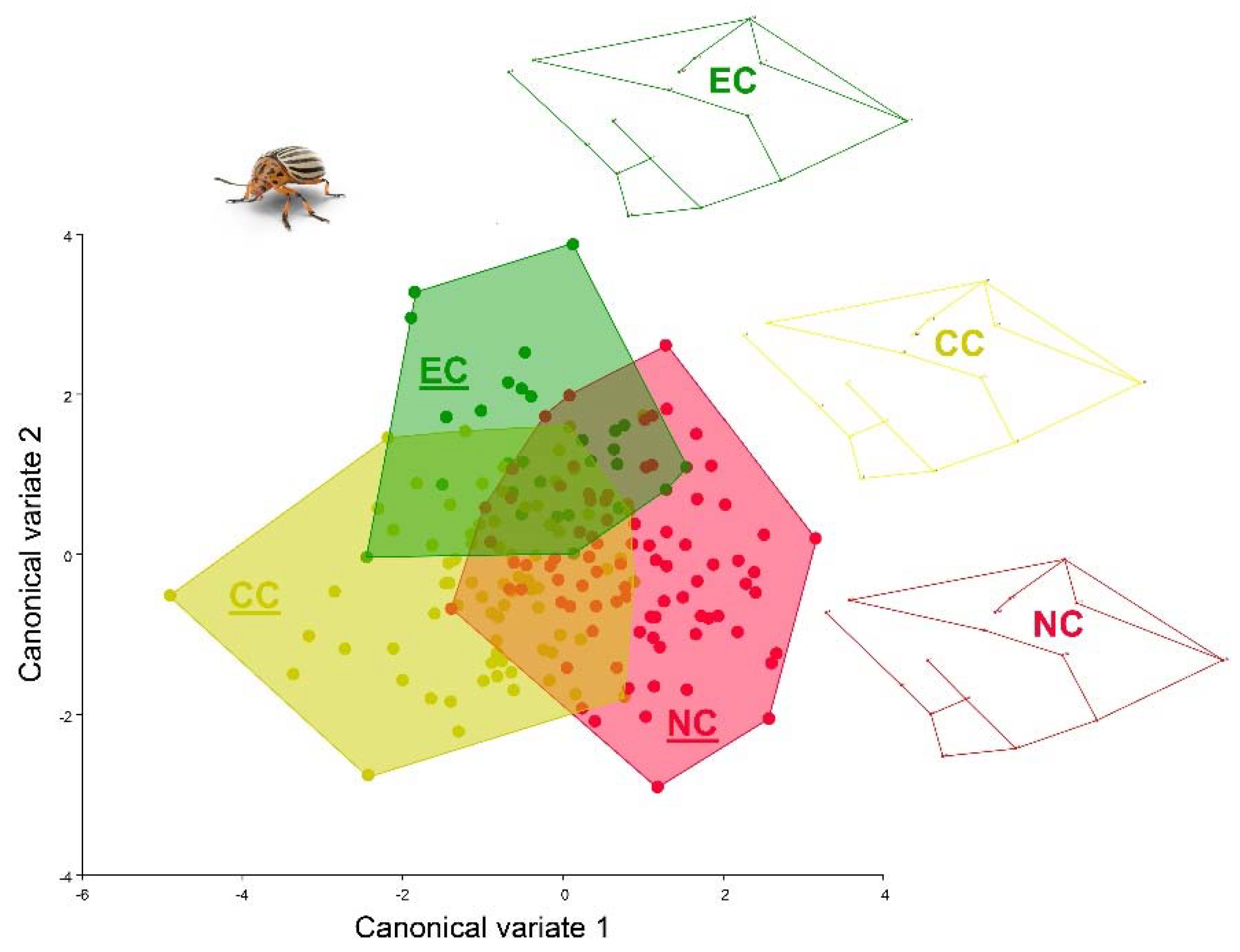
3.3. Geometric Morphometrics Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alyokhin, A.; Baker, M.; Mota-Sanchez, D.; Dively, G.; Grafius, E. Colorado potato beetle resistance to insecticides. Am. J. Potato Res. 2008, 85, 395–413. [Google Scholar] [CrossRef]
- Maceljski, M. Krumpirova zlatica (Leptinotarsa decemlineata Say). In Poljoprivredna Entomologija, 2nd ed.; Zrinski: Čakovec, Croatia, 2002; pp. 216–222. [Google Scholar]
- Ferro, D.N.; Logan, J.A.; Voss, R.H.; Elkinton, J.S. Colorado potato beetle (Coleoptera: Chrysomelidae) temperature-dependent growth and feeding rates. Environ. Entomol. 1985, 14, 343–348. [Google Scholar] [CrossRef]
- Alyokhin, A. Colorado potato beetle management on potatoes: Current challenges and future prospects. Fruit Veg. Cereal Sci. Biotechnol. 2009, 3, 10–19. [Google Scholar]
- Casagrande, R.A. The Colorado potato beetle: 125 years of mismanagement. Bull. Entomol. Soc. Am. 1987, 33, 142–150. [Google Scholar] [CrossRef]
- Gauthier, N.L.; Hofmaster, R.N.; Semel, M. History of Colorado potato beetle control. Adv. Potato Pest Manag. 1981, 23, 13–33. [Google Scholar]
- Arthropod Pesticide Resistance Database (APRD). Leptinotarsa decemlineata-Shown Resistance to Active Ingredient(s). Available online: https://www.pesticideresistance.org/display.php?page=species&arId=141 (accessed on 26 March 2022).
- Weber, D. Colorado beetle: Pest on the move. Pestic. Outlook 2003, 14, 256–259. [Google Scholar] [CrossRef]
- Cong, W.A.N.G.; Han, X.U.; Pan, X.B. Management of Colorado potato beetle in invasive frontier areas. J. Integr. Agric. 2020, 19, 360–366. [Google Scholar]
- Bradshaw, A.D. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar]
- Schlichting, C.D. The evolution of phenotypic plasticity. Ann. Rev. Ecol. Syst. 1986, 17, 667–693. [Google Scholar] [CrossRef]
- Berrigan, D.M.; Scheiner, S.M. Modelling the evolution of phenotypic plasticity. In Phenotypic Plasticity: Functional and Conceptual Approaches, 1st ed.; De Witt, T.J., Scheiner, S.M., Eds.; Oxford University Press: Oxford, UK, 2004; pp. 82–97. [Google Scholar]
- Van Kleunen, M.; Fisher, M. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol. 2005, 166, 49–60. [Google Scholar] [CrossRef]
- Helmuth, B.; Kingsolver, J.G.; Carrington, E. Biophysics, physiological ecology, and climate change: Does mechanism matter? Annu. Rev. Physiol. 2005, 67, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Schlichting, C.D. The role of phenotypic plasticity in diversification. In Phenotypic Plasticity: Functional and Conceptual Approaches, 1st ed.; de Witt, T.J., Scheiner, S.M., Eds.; Oxford University Press: Oxford, UK, 2004; pp. 191–200. [Google Scholar]
- Murren, C.J.; Denning, W.; Pigliucci, M. Relationships between vegetative and life history traits and fitness in a novel field environment: Impacts of herbivores. Evol. Ecol. 2005, 19, 583–601. [Google Scholar] [CrossRef]
- Cingel, A.; Savić, J.; Lazarević, J.; Ćosić, T.; Raspor, M.; Smigocki, A.; Ninković, S. Extraordinary adaptive plasticity of Colorado potato beetle: “Ten-Striped Spearman” in the era of biotechnological warfare. Int. J. Mol. Sci. 2016, 17, 1538. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Boiteau, G.; Alyokhin, A.; Ferro, D.N. The Colorado potato beetle in movement. Can. Entomol. 2003, 135, 1–22. [Google Scholar] [CrossRef]
- Estoup, A.; Guillemaud, T. Reconstructing routes of invasion using genetic data: Why, how and so what? Mol. Ecol. 2010, 19, 4113–4130. [Google Scholar] [CrossRef]
- Cristescu, M.E. Genetic reconstructions of invasion history. Mol. Ecol. 2015, 24, 2212–2225. [Google Scholar] [CrossRef]
- Grapputo, A.; Boman, S.; Lindstroem, L.; Lyytinen, A.; Mappes, J. The voyage of an invasive species across continents: Genetic variability of North American and European Colorado potato beetle populations. Mol. Ecol. 2005, 14, 4207–4219. [Google Scholar] [CrossRef]
- Jacobson, J.W.; Hsiao, T.H. Isozyme variation between geographic populations of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Ann. Entomol. Soc. Am. 1983, 76, 162–166. [Google Scholar] [CrossRef]
- Azeredo-Espin, A.M.L.; Schroder, R.F.W.; Huettel, M.D.; Sheppard, W.S. Mitochondrial DNA variation in geographic populations of Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera; Chrysomelidae). Experientia 1991, 47, 483–485. [Google Scholar] [CrossRef]
- Zehnder, G.W.; Sandall, L.; Tisler, A.M.; Powers, T.O. Mitochondrial DNA diversity among 17 geographic populations of Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Ann. Entomol. Soc. Am. 1992, 85, 234–240. [Google Scholar] [CrossRef]
- Azeredo-Espin, A.M.L.; Schroder, R.F.W.; Roderick, G.K.; Sheppard, W.S. Intraspecific mitochondrial DNA variation in the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Biochem. Genet. 1996, 34, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, A.P.; Berezovska, O.P. Genetic structure of populations of the Colorado potato beetle Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Russ. J. Genet. 2002, 38, 1256–1261. [Google Scholar] [CrossRef]
- Grapputo, A. Development and characterization of microsatellite markers in the Colorado potato beetle, Leptinotarsa decemlineata. Mol. Ecol. Notes 2006, 6, 1177–1179. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yang, J.; Li, Y.C.; Liu, N.; Zhang, R.Z. Genetic relationships of introduced Colorado potato beetle Leptinotarsa decemlineata populations in Xinjiang, China. Insect Sci. 2013, 20, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Przybylska, A.; Budziszewska, M.; Klejdysz, T.; Nawrot, J.; Obrępalska-Stęplowska, A. High stability of a mitochondrial genetic marker mtCOII in Polish Colorado potato beetle populations. Am. J. Potato Res. 2014, 91, 720–725. [Google Scholar] [CrossRef][Green Version]
- Izzo, V.M.; Chen, Y.H.; Schoville, S.D.; Wang, C.; Hawthorne, D.J. Origin of pest lineages of the Colorado potato beetle (Coleoptera: Chrysomelidae). J. Econ. Entomol. 2018, 111, 868–878. [Google Scholar] [CrossRef]
- Yang, F.Y.; Guo, J.J.; Liu, N.; Zhang, R.Z. Genetic structure of the invasive Colorado potato beetle Leptinotarsa decemlineata populations in China. J. Integr. Agric. 2020, 19, 350–359. [Google Scholar] [CrossRef]
- Jarne, P.; Lagoda, P.J. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 1996, 11, 424–429. [Google Scholar] [CrossRef]
- Barker, G.C. Microsatellite DNA: A tool for population genetic analysis. Trans. R. Soc. Trop. Med. Hyg. 2002, 96, S21–S24. [Google Scholar] [CrossRef]
- Özkan Koca, A.; Berkcan, S.B.; Laçın Alas, B.; Kandemir, İ. Population structure and pattern of geographic differentiation of Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) in Turkey. Pest Manag. Sci. 2022, 78, 3804–3814. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.S.; Chen, Y.H.; Groves, R.L.; Schoville, S.D. Landscape genomics of Colorado potato beetle provides evidence of polygenic adaptation to insecticides. Mol. Ecol. 2017, 26, 6284–6300. [Google Scholar] [CrossRef] [PubMed]
- Schoville, S.D.; Chen, Y.H.; Andersson, M.N.; Benoit, J.B.; Bhandari, A.; Bowsher, J.H.; Brevik, K.; Cappelle, K.; Chen, M.-J.M.; Childers, A.K.; et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 1931. [Google Scholar] [CrossRef]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef] [PubMed]
- Nantoume, A.D.; Andersen, S.B.; Jensen, B.D. Genetic differentiation of watermelon landrace types in Mali revealed by microsatellite (SSR) markers. Genet. Resour. Crop Evol. 2013, 60, 2129–2141. [Google Scholar] [CrossRef]
- Adams, D.; Rohlf, F.J.; Slice, D. A field comes of age: Geometric morphometrics in the 21st century. Hystrix Ital. J. Mammal. 2013, 24, 7–14. [Google Scholar]
- Klingenberg, C.P. Visualizations in geometric morphometrics: How to read and how to make graphs showing shape changes. Hystrix Ital. J. Mammal. 2013, 24, 15–24. [Google Scholar]
- Benítez, H.A.; Püschel, T.A. Modelando la varianza de la forma: Morfometría geométrica aplicaciones en biología evolutiva. Int. J. Morphol. 2014, 32, 998–1008. [Google Scholar] [CrossRef]
- Levine, E.; Oloumi-Sadeghi, H. Western corn rootworm (Coleoptera, Chrysomelidae) larval injury to corn grown for seed production following soybeans grown for seed production. J. Econ. Entomol. 1996, 89, 1010–1016. [Google Scholar] [CrossRef]
- Bouyer, J.; Ravel, S.; Dujardin, J.P.; De Meeüs, T.; Vial, L.; Thévenon, S.; Guerrini, L.; Sidibé, I.; Solano, P. Population structuring of Glossina palpalis gambiensis (Diptera: Glossinidae) according to landscape fragmentation in the Mouhoun river, Burkina Faso. J. Med. Entomol. 2007, 44, 788–795. [Google Scholar] [CrossRef]
- Benítez, H.A.; Lemic, D.; Bažok, R.; Gallardo-Araya, C.M.; Mikac, K.M. Evolutionary directional asymmetry and shape variation in Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae): An example using hind wings. Biol. J. Linn. Soc. 2014, 111, 110–118. [Google Scholar] [CrossRef]
- Lemic, D.; Benítez, H.A.; Püschel, T.A.; Gašparić, H.V.; Šatvar, M.; Bažok, R. Ecological morphology of the sugar beet weevil Croatian populations: Evaluating the role of environmental conditions on body shape. Zool. Anz. A. J. Comp. Zool. 2016, 260, 25–32. [Google Scholar] [CrossRef]
- Benítez, H.A.; Lemic, D.; Püschel, T.A.; Gašparić, H.V.; Kos, T.; Barić, B.; Bažok, R.; Živković, I.P. Fluctuating asymmetry indicates levels of disturbance between agricultural productions: An example in Croatian population of Pterostichus melas melas (Coleptera: Carabidae). Zool. Anz. 2018, 276, 42–49. [Google Scholar] [CrossRef]
- Lemic, D.; Benítez, H.A.; Bjeliš, M.; Órdenes-Claveria, R.; Ninčević, P.; Mikac, K.M.; Živković, I.P. Agroecological effect and sexual shape dimorphism in medfly Ceratitis capitata (Diptera: Tephritidae) an example in Croatian populations. Zool. Anz. 2020, 288, 118–124. [Google Scholar] [CrossRef]
- Pajač Živković, I.; Lemic, D.; Mešić, A.; Barić, B.; Órdenes, R.; Benítez, H.A. Effect of fruit host on wing morphology in Drosophila suzukii (Diptera: Drosophilidae): A first view using geometric morphometrics. Entomol. Res. 2018, 48, 262–268. [Google Scholar] [CrossRef]
- Lemic, D.; Bjeliš, M.; Ninčević, P.; Živković, I.P.; Popović, L.; Gašparić, H.V.; Benítez, H.A. Medfly phenotypic plasticity as a prerequisite for invasiveness and adaptation. Sustainability 2021, 13, 12510. [Google Scholar] [CrossRef]
- Lemic, D.; Mikac, K.M.; Kozina, A.; Benítez, H.A.; McLean, C.M.; Bažok, R. Monitoring techniques of the western corn rootworm are the precursor to effective IPM strategies. Pest Manag. Sci. 2016, 72, 405–417. [Google Scholar] [CrossRef]
- Živković, I.P.; Benitez, H.A.; Barić, B.; Drmić, Z.; Balaško, M.K.; Lemic, D.; Davila, J.H.D.; Mikac, K.M.; Bažok, R. Codling moth wing morphology changes due to insecticide resistance. Insects 2019, 10, 310. [Google Scholar] [CrossRef]
- Mikac, K.M.; Lemic, D.; Benítez, H.A.; Bažok, R. Changes in corn rootworm wing morphology are related to resistance development. J. Pest Sci. 2019, 92, 443–451. [Google Scholar] [CrossRef]
- Hood, C.S. Geometric morphometric approaches to the study of sexual size dimorphism in mammals. Hystrix 2000, 11, 77–90. [Google Scholar]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics, 1st ed.; Pompanon, F., Bonin, A., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 67–89. [Google Scholar]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high–performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3–1.new tools for the analysis of genome–wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE harvester. A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Rohlf, F.J. The tps series of software. Hystrix 2015, 26, 9. [Google Scholar]
- Rohlf, F.J.; Slice, D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Biol. 1990, 39, 40–59. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations FAO STAT. Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 31 May 2022).
- Garnier-Géré, P.; Chikhi, L. Population subdivision, Hardy–Weinberg equilibrium and the Wahlund effect. In eLS; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Puillandre, N.; Dupas, S.; Dangles, O.; Zeddam, J.L.; Capdevielle-Dulac, C.; Barbin, K.; Torres-Leguizamon, M.; Silvain, J.F. Genetic bottleneck in invasive species: The potato tuber moth adds to the list. Biol. Invasions 2008, 10, 319–333. [Google Scholar] [CrossRef]
- Mikac, K.M.; Lemic, D.; Bažok, R.; Benítez, H.A. Wing shape changes: A morphological view of the Diabrotica virgifera virgifera European invasion. Biol. Invasions 2016, 18, 3401–3407. [Google Scholar] [CrossRef]
- Karsten, M.; Addison, P.; Jansen van Vuuren, B.; Terblanche, J. Investigating population differentiation in a major African agricultural pest: Evidence from geometric morphometrics and connectivity suggests high invasion potential. Mol. Ecol. 2016, 25, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Garnier, S.; Magniez-Jannin, F.; Rasplus, J.Y.; Alibert, P. When morphometry meets genetics: Inferring the phylogeography of Carabus solieri using Fourier analyses of pronotum and male genitalia. J. Evol. Biol. 2005, 18, 269–280. [Google Scholar] [CrossRef]
- Camara, M.; Caro-Riano, H.; Ravel, S.; Dujardin, J.P.; Hervouet, J.P.; De MeEüs, T.; Bouyer, J.; Solano, P. Genetic and morphometric evidence for population isolation of Glossina palpalis gambiensis (Diptera: Glossinidae) on the Loos islands, Guinea. J. Med. Entomol. 2006, 43, 853–860. [Google Scholar] [CrossRef]
- Henriques, D.; Chávez-Galarza, J.; Teixeira, J.S.G.; Ferreira, H.; Neves, C.J.; Francoy, T.M.; Pinto, M.A. Wing geometric morphometrics of workers and drones and single nucleotide polymorphisms provide similar genetic structure in the Iberian honey bee (Apis mellifera iberiensis). Insects 2020, 11, 89. [Google Scholar] [CrossRef]
- Mikac, K.M.; Douglas, J.; Spencer, J.L. Wing shape and size of the western corn rootworm (Coleoptera: Chrysomelidae) is related to sex and resistance to soybean-maize crop rotation. J. Econ. Entomol. 2013, 106, 1517–1524. [Google Scholar] [CrossRef]
- Lemic, D.; Benítez, H.A.; Bažok, R. Intercontinental effect on sexual shape dimorphism and allometric relationships in the beetle pest Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae). Zool. Anz. A J. Comp. Zool. 2014, 253, 203–206. [Google Scholar] [CrossRef]
- Kadoić Balaško, M.; Mikac, K.M.; Benítez, H.A.; Bažok, R.; Lemic, D. Genetic and morphological approach for western corn rootworm resistance management. Agriculture 2021, 11, 585. [Google Scholar] [CrossRef]
- Voss, R.H.; Ferro, D.N. Phenology of flight and walking by Colorado potato beetle (Coleoptera: Chrysomelidae) adults in western Massachusetts. Environ. Entomol. 1990, 19, 117–122. [Google Scholar] [CrossRef]
- Lemic, D.; Mikac, K.M.; Ivkosic, S.A.; Bažok, R. The temporal and spatial invasion genetics of the western corn rootworm (Coleoptera: Chrysomelidae) in southern Europe. PLoS ONE 2015, 10, e0138796. [Google Scholar] [CrossRef] [PubMed]
- Drmić, Z.; Tóth, M.; Lemić, D.; Grubišić, D.; Pospišil, M.; Bažok, R. Area-wide mass trapping by pheromone-based attractants for the control of sugar beet weevil (Bothynoderes punctiventris Germar, Coleoptera: Curculionidae). Pest Manag. Sci. 2017, 73, 2174–2183. [Google Scholar] [CrossRef]
- Vreysen, M.J.B.; Robinson, A.S.; Hendrichs, J. Areawide control of insect pests. From research to field implementation. In Principles and Practice in Area-Wide Integrated Pest Management, Sterile Insect Technique, 1st ed.; Dyck, V.A., Hendrichs, J., Robinson, A.S., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 351–353. [Google Scholar]
- Furlan, L.; Chiarini, F.; Contiero, B.; Benvegnù, I.; Horgan, F.G.; Kos, T.; Lemic, D.; Bažok, R. Risk Assessment and Area-Wide Crop Rotation to Keep Western Corn Rootworm below Damage Thresholds and Avoid Insecticide Use in European Maize Production. Insects 2022, 13, 415. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kao, H.; Dong, S. Population genetic structure and gene flow of rare and endangered Tetraena mongolica Maxim. revealed by reduced representation sequencing. BMC Plant Biol. 2020, 20, 391. [Google Scholar]
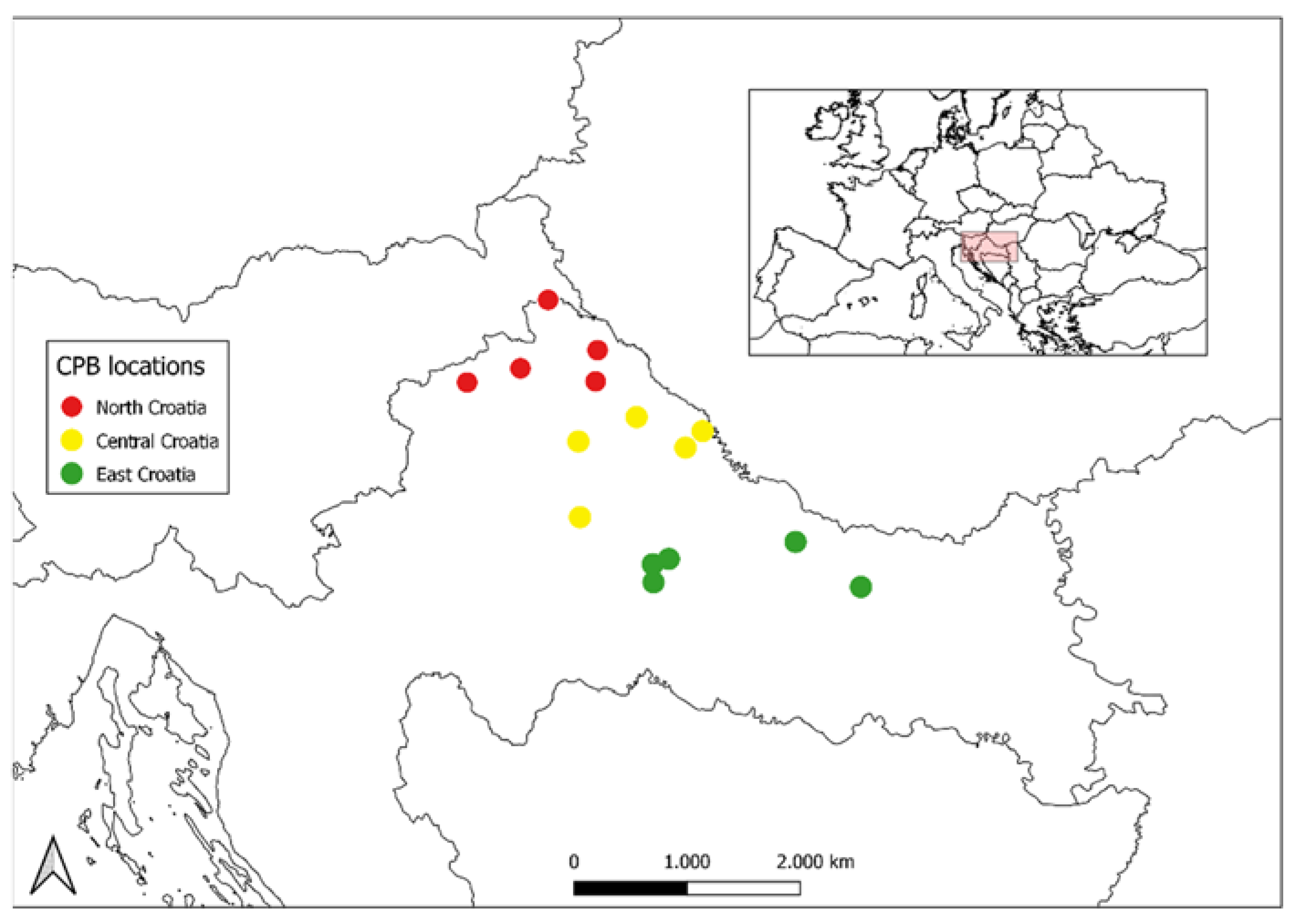



| Region | Population | Location | Lat. | Long. | n | CT |
|---|---|---|---|---|---|---|
| North Croatia | SVAM | Sv. Martin na Muri | 46°31′ | 16°21′ | 6 | 2018 |
| CEHO | Čehovec | 46°21′ | 16°37′ | 5 | 2019 | |
| VIDO | Vidovec | 46°17′ | 16°14′ | 5 | 2017 | |
| LUDB | Ludbreg | 46°15′ | 16°36′ | 5 | 2018 | |
| BEDN | Bednjs | 46°13′ | 15°58′ | 5 | 2018 | |
| Central Croatia | MLAD | Mladine | 46°02′ | 16°32′ | 6 | 2017 |
| STAR | Starigrad | 46°08′ | 16°49′ | 5 | 2017 | |
| DURD | Đurđevac | 46°02′ | 17°04′ | 6 | 2017 | |
| NVIR | Novo Virje | 46°05′ | 17°09′ | 6 | 2018 | |
| DRAG | Dragičevci | 45°47′ | 16°34′ | 5 | 2019 | |
| East Croatia | PASI | Pašijan | 45°38′ | 16°56′ | 6 | 2017 |
| GARE | Garešnica | 45°34′ | 16°56′ | 4 | 2017 | |
| HERC | Hercegovac | 45°39′ | 17°00′ | 6 | 2017 | |
| ZDEN | Zdenci | 45°34′ | 17°57′ | 5 | 2018 | |
| DMEL | Donji Meljani | 45°43′ | 17°37′ | 5 | 2017 |
| Region | n | A | P | Ho | He | FIS |
|---|---|---|---|---|---|---|
| North Croatia | 27 | 12,478 | 120 | 0.251 | 0.320 | 0.216 |
| Central Croatia | 28 | 12,539 | 193 | 0.258 | 0.323 | 0.201 |
| East Croatia | 27 | 12,503 | 150 | 0.255 | 0.326 | 0.218 |
| Source of Variation | SS | df | MS | F | p-Value | F crit |
|---|---|---|---|---|---|---|
| Between Groups | 0.008679583 | 15 | 0.000579 | 2.311803 | 0.004461 | 1.711235 |
| Within Groups | 0.056066667 | 224 | 0.00025 | |||
| Total | 0.06474625 | 239 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadoić Balaško, M.; Bažok, R.; Mikac, K.M.; Benítez, H.A.; Correa, M.; Lemic, D. Assessing the Population Structure of Colorado Potato Beetle Populations in Croatia Using Genetic and Geometric Morphometric Tools. Agronomy 2022, 12, 2361. https://doi.org/10.3390/agronomy12102361
Kadoić Balaško M, Bažok R, Mikac KM, Benítez HA, Correa M, Lemic D. Assessing the Population Structure of Colorado Potato Beetle Populations in Croatia Using Genetic and Geometric Morphometric Tools. Agronomy. 2022; 12(10):2361. https://doi.org/10.3390/agronomy12102361
Chicago/Turabian StyleKadoić Balaško, Martina, Renata Bažok, Katarina M. Mikac, Hugo A. Benítez, Margarita Correa, and Darija Lemic. 2022. "Assessing the Population Structure of Colorado Potato Beetle Populations in Croatia Using Genetic and Geometric Morphometric Tools" Agronomy 12, no. 10: 2361. https://doi.org/10.3390/agronomy12102361
APA StyleKadoić Balaško, M., Bažok, R., Mikac, K. M., Benítez, H. A., Correa, M., & Lemic, D. (2022). Assessing the Population Structure of Colorado Potato Beetle Populations in Croatia Using Genetic and Geometric Morphometric Tools. Agronomy, 12(10), 2361. https://doi.org/10.3390/agronomy12102361










