Growth Performance and Nitrogen Fixing Efficiency of Faba Bean (Vicia faba L.) Genotypes in Symbiosis with Rhizobia under Combined Salinity and Hypoxia Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Soils
2.2. Plant Materials and Experimental Plan
2.3. Growth Analysis
2.4. Plant Biomass
2.5. Chlorophyll Content and Gas Exchange Measurement
2.6. Photosynthetic Parameters
2.7. The Electrolyte Leakage
2.8. Lipid Peroxidation Assay
2.9. Proteins Assay
2.10. Sugar Assay Determination
2.11. Isolation of Bacterial Strains
2.12. DNA Isolation
2.13. Amplification of the nodC Gene
2.14. PCR Amplification of the 16S rRNA Gene
2.15. Nodulation Assay
2.16. Acetylene Reduction Assay (ARA)
2.17. Analysis of Bacteria Salinity Stress Tolerance
2.18. Statistical Analysis
3. Results
3.1. Vegetative Growth Analysis
3.2. Photosynthesis and Chlorophyll Contents
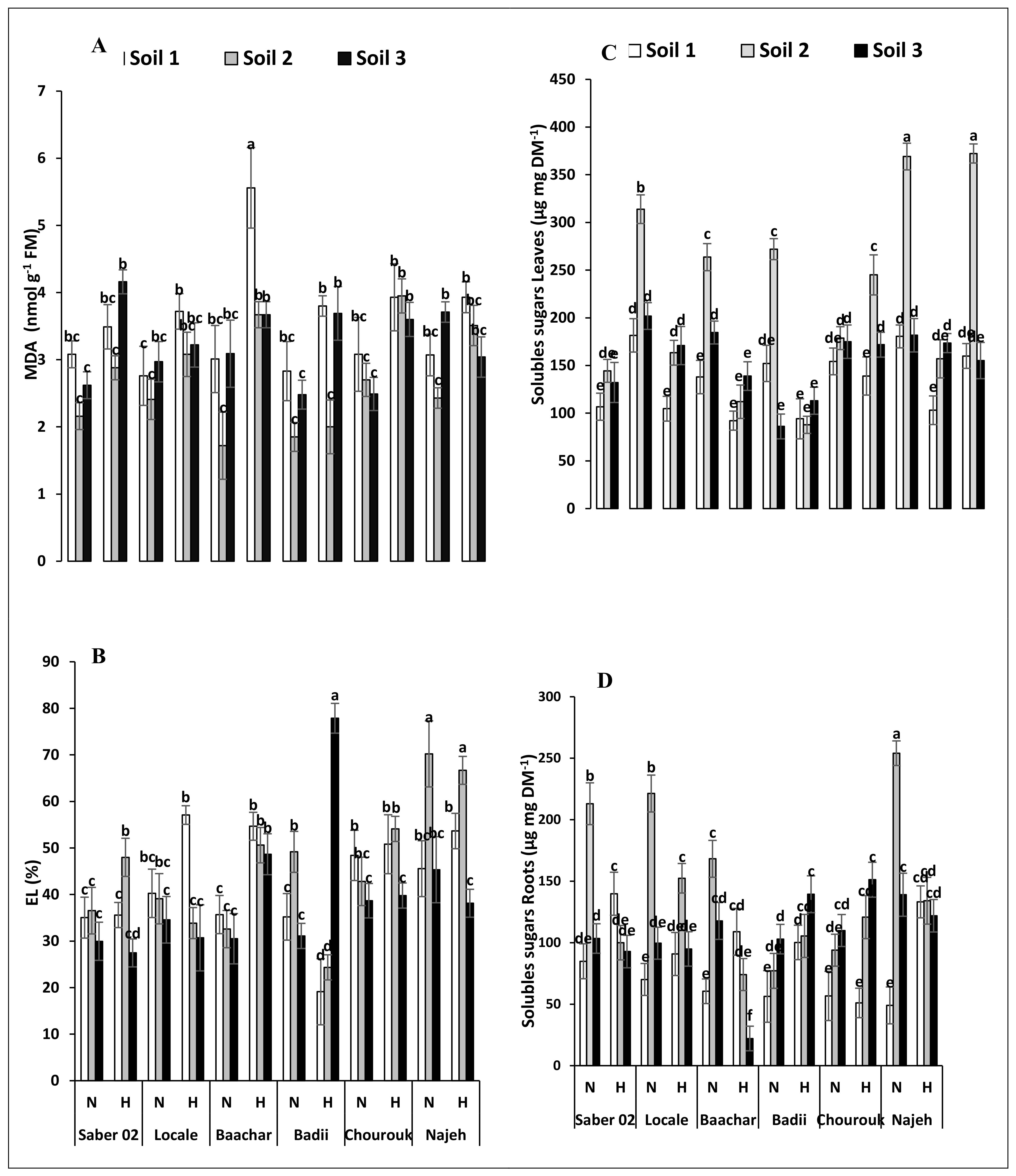
3.3. Peroxidation and Electrolyte Leakage
3.4. Carbohydrate Contents
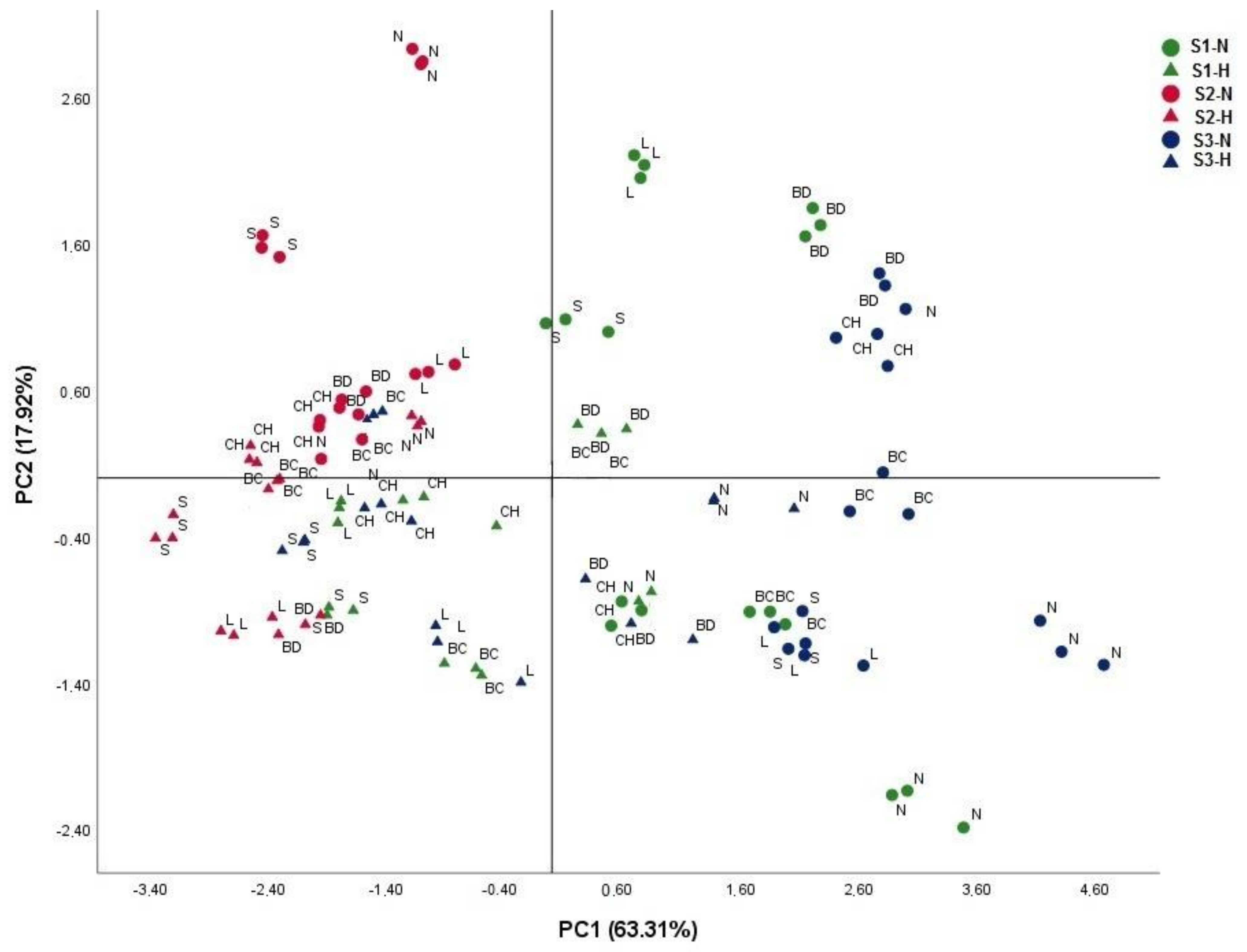
3.5. Selection of Efficient Genotype under Combined Salinity and Flooding
3.6. Rhizobial Strains Selection and nodC Amplification
3.7. Symbiotic Nitrogen Fixation Capacity
4. Discussions
4.1. Stress Conditions Differently Affect Genotype Productivity
4.2. Photosynthesis Does Not Limit Faba Bean Growth under Stress Conditions
4.3. Identification of Salt Tolerant Rhizobial Strains and Their Effect on Symbiosis Performance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, G.Q.; Ma, B.L.; Ren, C.Z. Growth, gas exchange, chlorophyll fluorescence, and ion content of naked oat in response to salinity. Crop Sci. 2007, 47, 123–131. [Google Scholar] [CrossRef]
- Bruning, B.; Rozema, J. Symbiotic nitrogen fixation in legumes: Perspectives for saline agriculture. Environ. Exp. Bot. 2013, 92, 134–143. [Google Scholar] [CrossRef]
- Glenn, E.P.; Brown, J.J.; Blumwald, E. Salt Tolerance and Crop Potential of Halophytes. Crit. Rev. Plant Sci. 1999, 18, 227–255. [Google Scholar] [CrossRef]
- Hafsi, C.; Romero-Puertas, M.C.; Gupta, D.K.; del Rio, L.A.; Sandalio, L.M.; Abdelly, C. Moderate salinity enhances the antioxidative response in the halophyte Hordeum maritimum L. under potassium deficiency. Environ. Exp. Bot. 2010, 69, 129–136. [Google Scholar] [CrossRef]
- Hachicha, M.; Job, J.O.; Mtimet, A. Les sols salés et la salinisation en Tunisie. Sols Tunis 1994, 15, 271–341. [Google Scholar]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2009, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Degl’Innocenti, E.; Hafsi, C.; Guidi, L.; Navari-Izzo, F. The effect of salinity on photosynthetic activity in potassium deficient barley species. J. Plant Physiol. 2009, 166, 1968–1981. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Blom, C.W.P.M.; Voeseneck, L.A.C.J. Flooding: The survival strategy of plants. Trends Ecol. Evol. 1996, 11, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, W. Aeration in higher plants. In Advances in Botanical Research; Woolhouse, H.W., Ed.; Academic Press: Cambridge, MA, USA, 1979; pp. 225–332. [Google Scholar]
- Voesenek, L.A.C.J.; Sasidharan, R. Ethylene-and oxygen signaling-drive plant survival during flooding. Plant Biol. 2013, 15, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Colmer, T.D. Plant tolerance of flooding stress recent advances. Plant Cell Environ. 2014, 37, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Striker, G.G.; Colmer, T.D. Flooding tolerance of forage legumes. J. Exp. Bot. 2017, 68, 1851–1872. [Google Scholar] [CrossRef] [Green Version]
- Qadir, M.; Oster, J. Crop and irrigation management strategies for saline-sodic soils and waters aimed at environmentally sustainable agriculture. Sci. Total Environ. 2004, 323, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Barrett-Lennard, E.G. The interaction between waterlogging and salinity in higher plants: Causes, consequences and implications. Plant Soil 2003, 253, 35–54. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Horchani, F.; Hajri, R.; Khayati, H.; Aschi-Smiti, S. Physiological responses of tomato plants to the combined effect of root hypoxia and NaCl salinity. J. Phytol. 2010, 2, 36–46. [Google Scholar]
- Gorai, M.; Ennajeh, M.; Khemira, H.; Neffati, M. Combined effect of NaCl-salinity and hypoxia on growth, photosynthesis, water relations and solute accumulation in Phragmites australis plants. Flora 2010, 205, 462–470. [Google Scholar] [CrossRef]
- Striker, G.G.; Teakle, N.L.; Colmer, T.D.; Barrett-Lennard, E.G. Growth responses of Melilotus siculus accessions to combined salinity and root-zone hypoxia are correlated with differences in tissue ion concentrations and not differences in root aeration. Environ. Exp. Bot. 2015, 109, 89–98. [Google Scholar] [CrossRef]
- Munir, R.; Konnerup, D.; Khan, H.A.; Siddique, K.H.M.; Colmer, T.D. Sensitivity of chickpea and faba bean to root-zone hypoxia, elevated ethylene, and carbone dioxide. Plant Cell Environ. 2019, 42, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Achour, A.; Benjedou, H.; Kadiri, A.; Fares, I.; Belkhodja, M. Vicia faba (L.) physiological and biochemical response under saline conditions. Int. J. Agric. Biosci. 2020, 9, 36–42. [Google Scholar]
- Yang, R.; Yin, Y.; Guo, L.; Han, Y.; Gu, Z. Sequence analysis of diamine oxidase gene from faba bean and its expression related to g-aminobutyric acid accumulation in seeds germinating under hypoxia-NaCl stress. J. Sci. Food Agric. 2014, 94, 1585–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wang, S.; Yin, Y.; Gu, Z. Hypoxia treatment on germinating faba bean (Vicia faba L.) seeds enhances GABA-related protection against salt stress. Chil. J. Agric. Res. 2015, 75, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Mnasri, B.; Mrabet, M.; Laguerre, G.; Aouani, M.E.; Mhamdi, R. Salt-tolerant rhizobia isolated from a Tunisian oasis that are highly effective for symbiotic N2-fixation with Phaseolus vulgaris constitute a novel biovar (bv. mediterranense) of Sinorhizobium meliloti. Arch. Microbiol. 2007, 187, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Tajini, F.; Drevon, J.; Lamouchi, L.; Aouani, M.E.; Trabelsi, M. Response of common bean lines to inoculation: Comparison between the Rhizobium tropici CIAT899 and the native Rhizobium etli 12a3 and their persistence in Tunisian soils. World J. Microbiol. Biotechnol. 2008, 24, 407–417. [Google Scholar] [CrossRef]
- Mhadhbi, H.; Fotopoulos, V.; Mylona, P.V.; Jebara, M.; Polidoros, A.N.; Aouani, M.E. Role of antioxidant gene-enzyme responses in Medicago truncatula genotypes with different degrees of sensitivity to high salinity. Physiol. Plant 2011, 141, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Institute of Soil Science, Chinese Academy of Sciences. Soil Physical and Chemical Analysis. Physical and Chemical Analyses of Soils; Shanghai Academic Press: Shanghai, China, 1980. [Google Scholar]
- Wintermans, J.; Mots, A. Spectrophotometric characteristic of chlorophylls a and b and their pheophytins in ethanol. Biochem. Biophys. Acta 1965, 109, 448–453. [Google Scholar]
- Fan, L.; Zheng, S.; Wang, X. Antisense suppression of phospholipase D alpha retards abscisic acid-and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 1997, 9, 2183–2196. [Google Scholar] [CrossRef]
- Cakmak, I.; Horst, W.J. Effect of aluminiumon lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol. Plant 1991, 83, 463–468. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Horchani, F.; Khayati, H.; Raymond, P.; Brouquisse, R.; Aschi-Smiti, S. Contrasted effects of prolonged root hypoxia on tomato (Solanum lycopersicum) roots and fruits metabolism. J. Agron. Crop Sci. 2009, 195, 313–318. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Handbook of Rhizobia: Methods in Legume Rhizobium Technology; Springer: Berlin, Germany, 1985. [Google Scholar]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria; Blackwell: Oxford, UK, 1970. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Smit, J.A.; Seidman, J.C.; Struhl, K.S. Current Protocols in Molecular Biology; Wiley: New York, NY, USA, 1994. [Google Scholar]
- Mhamdi, R.; Laguerre, G.; Aouani, M.E.; Mars, M.; Amarger, N. Different species and symbiotic genotypes of field rhizobia can nodulate Phaseolus vulgaris in Tunisian soils. FEMS Microbiol. Ecol. 2002, 41, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Moschetti, G.; Peluso, A.L.; Protopapa, A.; Anastasio, M.; Pepe, O.; Defez, R. Use of nodulation pattern, stress tolerance, nodC amplification, RAPD-PCR and RFLP-16S rDNA analysis to discriminate genotypes of Rhizobium leguminosarum biovar viciae. Syst. Appl. Microbiol. 2005, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Mhadhbi, H.; Jebara, M.; Limam, F.; Huguet, T.; Aouani, M.E. Interaction between Medicago truncatula lines and Sinorhizobium meliloti strains for symbiotic efficiency and nodule antioxidant activities. Physiol. Plant. 2005, 124, 4–11. [Google Scholar] [CrossRef]
- Vadez, V.; Rodier, F.; Payre, H.; Drevon, J.J. Nodule permeability to O2 and nitrogenase-linked respiration in bean genotypes varying in the tolerance of N2 fixation to P deficiency. Plant Physiol. Biochem. 1996, 34, 871–878. [Google Scholar]
- Hardy, R.W.F.; Bums, C.; Hebert, R.R.; Holsten, R.D. Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol. Biochem. 1973, 5, 47–81. [Google Scholar] [CrossRef]
- Mhadhbi, H.; Fotopoulos, V.; Djebali, N.; Polidoros, A.N.; Aouani, M.E. Behaviors of Medicago truncatula-Sinorhizobium meliloti symbioses under osmotic stress in relation with the symbiotic partner input: Effects on nodule functioning and protection. J. Agron. Crop. Sci. 2009, 195, 225–231. [Google Scholar] [CrossRef]
- Mrabet, M.; Zribi, K.; Mhadhbi, H.; Djébali, N.; Mhamdi, R.; Aouani, M.E.; Nakamura, K. Salt tolerance of a Sinorhizobium meliloti strain isolated from dry lands: Growth capacity and protein profile changes. Ann. Microbiol. 2011, 61, 361–369. [Google Scholar] [CrossRef]
- James, E.K.; Sprent, J.I. Development of N2-fixing nodules on the wetland legume Lotus uliginosus exposed to conditions of flooding. New Phytol. 1999, 142, 219–231. [Google Scholar] [CrossRef]
- Aschi-Smiti, S.; Chaibi, W.; Brouquisse, R.; Ricard, B.; Saglio, P. Assessment of enzyme induction and aerenchyma formation as mechanisms for flooding tolerance in Trifolium subterraneum ‘Park’. Ann. Bot. 2003, 91, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konnerup, D.; Toro, G.; Pedersen, O.; Colmer, T.D. Waterlogging tolerance, tissue nitrogen and oxygen transport in the forage legume Melilotus siculus: A comparison of nodulated and nitrate-fed plants. Ann. Bot. 2018, 121, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.J.; Ligero, F.; Lluch, C. Effects of salt stress on growth and nitrogen fixation by pea, faba-bean, common bean and soybean plants. Soil Biol. Biochem. 1994, 26, 371–376. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef] [Green Version]
- Velasco, N.F.; Ligarreto, G.A.; Diaz, H.R.; Fonseca, L.P.M. Photosynthetic responses and tolerance to root-zone hypoxia stress of five bean cultivars (Phaseolus vulgaris L.). S. Afr. J. Bot. 2019, 123, 200–207. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Jimenez, A.; Mullineaux, P.M.; Sevilla, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defenses. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Saha, P.; Chatterjee, P.; Biswas, A.K. NaCl pretreatment alleviates salt stress by enhancement of antioxidant defense system and osmolyte accumulation in mungbean (Vigna radiata L. Wilczek). Indian J. Exp. Biol. 2010, 48, 593–600. [Google Scholar]
- Mahlooji, M.; Seyed Sharifi, R.; Razmjoo, J.; Sabzalian, M.R.; Sedghi, M. Effect of salt stress on photosynthesis and physiological parameters of three contrasting barley genotypes. Photosynthetica 2018, 56, 549–556. [Google Scholar] [CrossRef]
- Rajhi, I.; Benmoussa, S.; Neji, I.; Baccouri, B.; Chikha, M.; Chammakhi, C.; Amri, M.; Mhadhbi, H. Photosynthetic and physiological responses of small seeded faba bean genotypes (Vicia faba L.) to salinity stress: Identification of a contrasting pair towards salinity. Photosynthetica 2020, 58, 174–185. [Google Scholar] [CrossRef]
- Tirichine, L.; de Billy, F.; Huguet, T. Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol. 2000, 123, 845–852. [Google Scholar] [CrossRef] [Green Version]
- Jebara, M.; Mhamdi, R.; Aouani, M.E.; Ghrir, R.; Mars, M. Genetic diversity of Sinorhizobium populations recovered from different Medicago varieties cultivated in Tunisian soils. Can. J. Microbiol. 2001, 47, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Mhadhbi, H.; Jebara, M.; Zitoun, A.; Limam, F.; Aouani, M.E. Symbiotic effectiveness and response to mannitol-mediated osmotic stress of various chickpea–rhizobia associations. World J. Microbiol. Biotechnol. 2008, 24, 027–1035. [Google Scholar] [CrossRef]
- Giller, K.E. Nitrogen Fixation in Tropical Cropping Systems; CAB International: Wallingford, UK, 2001; pp. 14–56. [Google Scholar]
- Elsheikh, E.A.E.; Wood, M. Nodulation and N2 fixation by soybean inoculated with salt-tolerant rhizobia or salt-sensitive brady rhizobia in saline soil. Soil Biol. Biochem. 1995, 27, 657–661. [Google Scholar] [CrossRef]
- Dardanelli, M.; Angelini, J.; Fabra, A. A calcium-dependent bacterial surface protein is involved in the attachment of rhizobia to peanut roots. Can. J. Microbiol. 2003, 49, 399–405. [Google Scholar] [CrossRef]
- Fraysse, N.; Couderc, F.; Poinsot, V. Surfacepolysaccharide involvement in establishing theRhizobium-legume symbiosis. Eur. J. Biochem. 2003, 270, 1365–1380. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Frugoli, J.A.; Smith, L.S.; Long, S.R.; Cook, D.R. Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 2003, 131, 998–1008. [Google Scholar] [CrossRef] [Green Version]
| Identification | Unit | Soils | ||
|---|---|---|---|---|
| Soil 1 | Soil 2 | Soil 3 | ||
| Conductivity | µs cm−1 | 153 | 1849 | 173 |
| Organic material | g kg−1 DM | 36.4 | 24.7 | 44.1 |
| Total organic carbon(TOC) | g kg−1 DM | 16.5 | 8.5 | 11.3 |
| Total nitrogen | g kg−1 DM | 2.1 | 0.9 | 1.4 |
| Total phosphorus | g kg−1 DM | 1.1 | 1.3 | 1.4 |
| Exchangeable Potassium | g kg−1 DM | 2.82 | 2.28 | 4.39 |
| Clay | % | 12.20 | 6.90 | 9.60 |
| Silt | % | 6.38 | 7.70 | 52.10 |
| Sand | % | 78.50 | 85.20 | 38 |
| Soils | Varieties | Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SDM [mg Plant−1] | RDM [mg Plant−1] | Shoots Length [cm] | Roots Length [cm] | Nodules Number [N plant−1] | |||||||
| N | H | N | H | N | H | N | H | N | H | ||
| Soil 1 | Saber | 3 d | 1.9 d | 0.7 cd | 0.4 e | 69 b | 66 b | 31 b | 11 d | 17 e | 5 fg |
| Locale | 2 d | 1.7 d | 1.1 c | 0.5 de | 66.3 b | 66.3 b | 38.3 a | 8.5 d | 19 e | 3 g | |
| Bachar | 3.3 c | 2.5 d | 2.1 b | 1.1 c | 71 b | 69 b | 26 b | 11.7 d | 43 b | 17 e | |
| Badii | 4 c | 2.2 d | 1.7 b | 1.3 bc | 71 b | 70 b | 32.7 ab | 20 c | 31 c | 25 d | |
| Chourouk | 3.2 c | 2.9 cd | 1.5 b | 1.2 bc | 65.3 b | 58 bc | 27.7 b | 11 d | 25 cd | 10 f | |
| Najeh | 5.2 bc | 4.8 bc | 2.7 a | 1.7 b | 62 b | 84.7 a | 33.3 b | 12.3 d | 54 a | 31 c | |
| Soil 2 | Saber | 0.9 e | 0.8 e | 0.5 de | 0.4 e | 42.3 d | 40 d | 15.5 cd | 4.3 e | 1 g | 0 |
| Locale | 1.2 e | 1.1 e | 1.1 c | 0.8 cd | 58 bc | 44.3 d | 27.7 b | 12 d | 0 | 0 | |
| Bachar | 1.1 e | 0.9 e | 0.9 cd | 0.6 de | 55.2 bc | 51.3 c | 18 c | 12.3 d | 1 g | 1 g | |
| Badii | 1.2 e | 1.1 e | 0.6 de | 0.6 de | 53.3 c | 50.3 c | 22.7 bc | 18 c | 4 g | 2 g | |
| Chourouk | 1.3 e | 1.2 e | 0.6 e | 0.4 e | 56.3 bc | 45.7 d | 17.7 c | 13.7 d | 2 g | 2 g | |
| Najeh | 1.6 e | 1.4 e | 0.5 e | 0.8 d | 59.7 bc | 63.3 b | 19.2 c | 23 bc | 5 g | 2 g | |
| Soil 3 | Saber | 7.6 a | 1.7 de | 1.6 b | 0.7 d | 67.3 b | 49 c | 32.7 b | 10.3 d | 26 cd | 6 fg |
| Locale | 4.5 c | 4.0 c | 2.0 b | 1.2 c | 76.7 a | 59.7 b | 26.3 b | 9.7 d | 28 c | 18 e | |
| Bachar | 6.8 ab | 1.7 de | 1.6 b | 0.4 e | 80.7 a | 65 b | 32.3 b | 11.3 d | 34 c | 9 f | |
| Badii | 5.7b | 5.5 b | 1.6 b | 1.1 c | 80.7 a | 59 bc | 41 a | 26.3 b | 27 cd | 27 cd | |
| Chourouk | 5.6b | 3.1 cd | 1.8 b | 0.8 cd | 79.7 a | 58 bc | 31.3 b | 5.3 e | 27 cd | 9 f | |
| Najeh | 7.7 a | 6.3 b | 2.5 a | 1.6 bc | 84.3 a | 67.7 b | 40 a | 19.7 c | 44 b | 30 c | |
| Soils | Varieties | Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A [µmol CO2 m−1 s−1] | Gs [mol m−1 s−1] | E [mmol H2O2 m−1 s−1] | Ci [µmol CO2 mol−1] | Chlorophyll [mg g−1 FM] | |||||||
| N | H | N | H | N | H | N | H | N | H | ||
| Soil 1 | Saber | 11.14 cd | 2.99 h | 0.07 g | 0.03 h | 0.74 fg | 0.95 f | 109 g | 234 de | 15.4 c | 3.8 e |
| Locale | 13.54 b | 10.44 d | 0.16 d | 0.13 e | 1.66 e | 2.93 d | 220 e | 236 de | 21.4 ab | 8.8 d | |
| Bachar | 10.06 d | 8.86 e | 0.08 fg | 0.03 h | 1.29 ef | 4.56 b | 184 f | 325 b | 10.1 cd | 4.4 de | |
| Badii | 13.95 b | 9 de | 0.12 e | 0.08 fg | 0.95 f | 2.60 d | ND | 248 d | 24 a | 14.4 c | |
| Chourouk | 11.17 cd | 6.40 f | 0.11 i | 0.06 g | 1.05 f | 2.49 d | 124 g | 251 d | 6.9 de | 11.7 c | |
| Najeh | 10.40 d | 4.49 g | 0.12 e | 0.09 f | 1.13 f | 1.67 e | 183 f | 221 e | 6 de | 9.6 cd | |
| Soil 2 | Saber | 4.89 g | 4.61 g | 0.11 ef | 0.03 h | 0.35 g | 0.95 f | 125 g | 321 b | 19 b | 8.3 d |
| Locale | 6.25 f | 2.09 hi | 0.07 g | 0.07 g | 0.54 g | 1.40 ef | 175 f | 338 b | 13 c | 4.4 e | |
| Bachar | 6.24 f | 1.36 i | 0.09 f | 0.07 g | 1.02 f | 1.46 ef | 186 f | 367 a | 10.8 cd | 9.6 d | |
| Badii | 5.85 f | 4.45 g | 0.13 e | 0.07 g | 0.82 f | 2.38 d | 289 c | 318 b | 11.8 c | 3 e | |
| Chourouk | 6.06 f | 3.42 h | 0.10 f | 0.08 fg | 1.18 f | 1.67 e | 284 c | 321 b | 11.1 cd | 10.4 cd | |
| Najeh | 4.37 g | 2.87 h | 0.06 g | 0.04 h | 0.64 fg | 1.49 ef | 230 de | 336 b | 25.9 a | 10.7 cd | |
| Soil 3 | Saber | 8.37 e | 5.27 fg | 0.19 c | 0.17 d | 4.28 b | 5.53 a | 291 c | 327 b | 8.5 d | 8.3 d |
| Locale | 12.31 c | 8.04 e | 0.14 de | 0.06 g | 2.40 d | 4.10b c | 216 e | 271 cd | 8.8 d | 8.6 d | |
| Bachar | 8.49 e | 8.46 e | 0.11 ef | 0.09 f | 3.17 cd | 3.31 c | 243 d | 250 d | 13.5 c | 12.8 c | |
| Badii | 9.54 d | 9.96 d | 0.16 d | 0.12 e | 4.08 bc | 4.45 b | 233 de | 275 c | 19.9 b | 8.2 d | |
| Chourouk | 10.07 d | 1.36 i | 0.12 e | 0.01 i | 1.35 i | 3.61 c | 217 e | 213 e | 19.8 b | 12 c | |
| Najeh | 9.58 de | 16.41 a | 0.11 ef | 0.22 b | 3.51 c | 5.98 a | 222 e | 229 de | 10.3 cd | 15.3 c | |
| Soil (S) | Genotypes (G) | Treatment (T) | S*G*T | |
|---|---|---|---|---|
| RL [cm plant−1] | 60.587 ** | 7.020 ** | 767.821 ** | 17.303 ** |
| RDM [mg plant−1] | 1070.191 ** | 186.526 ** | 692.104 ** | 89.325 ** |
| Number of Nod [N plant−1] | 364.947 ** | 12.171 ** | 322.520 ** | 21.657 ** |
| SL [cm plant−1] | 389.572 ** | 30.116 ** | 124.614 ** | 26.336 ** |
| SDM [mg Plant−1] | 967.465 ** | 78.886 ** | 344.263 ** | 38.445 ** |
| Chlorophyll [mg g−1 FM] | 626.226 ** | 1089.681 ** | 19,213.788 ** | 1771.654 ** |
| Strains Code | GenBank Accession Numbers | Identical Strains | % Similarity Compared to the Identified Sequence | Salinity Tolerance Limits (mM) | ARA (µmol h−1Plant−1) | Shoots Length (cm) | Roots Length (cm) | Nodules Number (NN/Plant-1) |
|---|---|---|---|---|---|---|---|---|
| 1 | MK809163 | KT387834.1 Rhizobium sp. L5 | 98.98 | 800 | 30.1 ± 0.47 | 29.7 ± 2.5 | 15.2 ± 1.3 | 36 ± 1 |
| 2 | MK809170 | MH345076.1 Rhizobium laguerreae strain PEPV08 | 98.69 | 350 | 22.8 ± 3.26 | 62 ± 1.7 | 37 ± 1.7 | 20 ± 3 |
| 3 | MK809176 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 98.65 | 350 | 14.7 ± 1.43 | 58.3 ± 1.5 | 35.7 ± 2.1 | 11 ± 3 |
| 7 | MK809181 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 97.92 | 400 | 5.6 ± 0.99 | 59 ± 1.7 | 44.6 ± 0.6 | 0 |
| 12 | MK809162 | EU074196.1 Rhizobium leguminosarum strain | 98.80 | 800 | 53.0 ± 1.42 | 81.3 ± 2.3 | 39.3 ± 1.2 | 67 ± 4 |
| 21 | MK809164 | EF549399.1 Rhizobium sp. CCBAU 83480 | 98.78 | 600 | 41.7 ± 1.92 | 69 ± 1 | 27 ± 3 | 50 ± 3 |
| 23 | MK809165 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 98.69 | 800 | 28.2 ± 0.97 | 74.3 ± 2.5 | 41 ± 1.7 | 31 ± 4 |
| 24 | MK809166 | KT387835.1 Rhizobium sp. L15 | 98.82 | 450 | 37.6 ± 0.35 | 66.6 ± 3.1 | 36.6 ± 3.1 | 43 ± 3 |
| 26 | MK809167 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 99.26 | 600 | 44.5 ± 1.08 | 62.6 ± 2.5 | 50.3 ± 1.5 | 56 ± 4 |
| 28 | MK809168 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 98.54 | 600 | 35.2 ± 0.57 | 66.3 ± 4 | 32.3 ± 2.5 | 47 ± 3 |
| 29 | MK809169 | MH345080.1 Rhizobium laguerreae strain PEPV31 | 98.97 | 500 | 6.7 ± 0.99 | 61.3 ± 2.3 | 37.6 ± 1.2 | 0 |
| 34 | MK809171 | KJ532459.1 Rhizobium sp. UFSM-B74 | 99.16 | 350 | 12.3 ± 1.42 | 63 ± 3.6 | 42.3 ± 2.5 | 10 ± 3 |
| 36 | MK809172 | MH345080.1 Rhizobium laguerreae strain PEPV31 | 92 | 350 | 10.8 ± 1.69 | 76.6 ± 1.5 | 34.3 ± 0.6 | 9 ± 3 |
| 37 | MK809173 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 98.68 | 350 | 12.3 ± 1.83 | 76.6 ± 2.5 | 31 ± 1 | 9 ± 3 |
| 38 | MK809174 | JN105993.1 Rhizobium leguminosarum strain 35-1 | 98.97 | 350 | 14.3 ± 0.84 | 72.3 ± 2.5 | 37.6 ± 2.5 | 11 ± 3 |
| 39 | MK809175 | MH345080.1 Rhizobium laguerreae strain PEPV31 | 97.36 | 600 | 24.7 ± 1.79 | 65.6 ± 3.8 | 24.3 ± 3.1 | 21 ± 1 |
| 41 | MK809177 | KX226363.1 Rhizobium leguminosarum strain PM25 | 95.26 | 800 | 30.5 ± 2.24 | 74 ± 2 | 25 ± 5 | 37 ± 2 |
| 42 | MK809178 | JF974177.1 Rhizobium sp. PZ-Ar-6-11 | 98.62 | 800 | 20.1 ± 2.27 | 70 ± 3 | 31.3 ± 1.2 | 26 ± 0 |
| 45 | MK809179 | KU878960.1 Rhizobium sp. SWF23008 | 97.05 | 600 | 25.4 ± 0.97 | 77.3 ± 2.5 | 32.3 ± 4.5 | 22 ± 0 |
| 48 | MK809180 | KJ634549.1 Rhizobium leguminosarum bv. trifolii strain SCD23-2 | 98.36 | 350 | 9.6 ± 1.08 | 66 ± 1.7 | 23.6 ± 2.5 | 5 ± 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benmoussa, S.; Nouairi, I.; Rajhi, I.; Rezgui, S.; Manai, K.; Taamali, W.; Abbes, Z.; Zribi, K.; Brouquisse, R.; Mhadhbi, H. Growth Performance and Nitrogen Fixing Efficiency of Faba Bean (Vicia faba L.) Genotypes in Symbiosis with Rhizobia under Combined Salinity and Hypoxia Stresses. Agronomy 2022, 12, 606. https://doi.org/10.3390/agronomy12030606
Benmoussa S, Nouairi I, Rajhi I, Rezgui S, Manai K, Taamali W, Abbes Z, Zribi K, Brouquisse R, Mhadhbi H. Growth Performance and Nitrogen Fixing Efficiency of Faba Bean (Vicia faba L.) Genotypes in Symbiosis with Rhizobia under Combined Salinity and Hypoxia Stresses. Agronomy. 2022; 12(3):606. https://doi.org/10.3390/agronomy12030606
Chicago/Turabian StyleBenmoussa, Sabrine, Issam Nouairi, Imen Rajhi, Saber Rezgui, Khediri Manai, Wael Taamali, Zouhaier Abbes, Kais Zribi, Renaud Brouquisse, and Haythem Mhadhbi. 2022. "Growth Performance and Nitrogen Fixing Efficiency of Faba Bean (Vicia faba L.) Genotypes in Symbiosis with Rhizobia under Combined Salinity and Hypoxia Stresses" Agronomy 12, no. 3: 606. https://doi.org/10.3390/agronomy12030606






