Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies
Abstract
:1. Introduction
2. Origin and Germplasm Resources
3. Traditional Breeding: Progress and Prospects
3.1. Challenges and Opportunities
3.2. Breeding for Yield and Plant Morphological Traits
3.3. Breeding for Resistance to Biotic Stresses
3.4. Breeding for Tolerance to Abiotic Stresses
3.4.1. Drought Tolerance
3.4.2. Soil Limiting Factors
3.4.3. High Temperature and Cold Injury
3.5. Breeding for Quality Traits
3.5.1. Human Consumption
3.5.2. Animal Nutrition
4. Molecular Breeding: Progress and Prospects
- (i) Improving the efficiency of selection: DNA-based markers and other genomic tools can be used to select for genes of importance more effectively and assess the existing genetic variation within the species to assist selecting parents for hybridization [81];
- (ii) Enhancing favourable gene action: Molecular tools can be used to understand the gene action and breeding value at various loci distributed across the genomes. Molecular cloning of QTLs can provide novel insights about the biology and nature of quantitative traits, and
- (iii) Expansion of useful genetic diversity for crop improvement: Molecular and recombinant DNA technology approaches can be used to identify and incorporate genes that are not otherwise accessible or available through crossing [82].
4.1. Marker Assisted Selection
4.1.1. Markers for Ascochyta Resistance
| Trait | Loci/QTL | Chromosome | Mapping populations | Linked markers | References |
|---|---|---|---|---|---|
| Rust resistance | Uvf1 | unknown | 2N52 × Vf176 (F2) | OPI20900/OPL181032 | [87] |
| Broomrape resistance | Oc1 | I | Vf6 × Vf136 (F2) | OPJ13686/OPAC02730 | [3] |
| Oc2 | VI | OPAC06342/OPN07849 | |||
| Oc3 | II | OPW15533/OPAA07807 | |||
| Oc2 | VI.B | Vf6 × Vf136 (RILs) | OPAI131018/OPAC06396 | [91] | |
| Oc3 | II.A | OPM15794/PisGEN 4_3_1 | |||
| Oc4 | I.A | OPAB01438/OPM181192 | |||
| Oc5 | I.A | OPM18620/OPA17524 | |||
| Oc2_C3 a | VI.B | Vf6 × Vf136 (RILs) | OPAG11/OPI5 | [92] | |
| Oc2_C4 | VI.B | OPAG11/OPI5 | |||
| Oc2_M4 | VI.B | OPAI131018/OPAC06396 | |||
| Oc3_C3 | II.A | OPM15794/Pis_GEN_4_3_1 | |||
| Oc3_C4 | II.A | OPM15794/Pis_GEN_4_3_1 | |||
| Oc4_C4 | I.A | OPB03289 | |||
| Oc5_M4 | I.A | OPM181620/OPA17524 | |||
| Oc6_C3 | I.A | OPG071714/OPH01900 | |||
| Oc7_C3 | II.A | OPP10 | |||
| Oc8_M4 | II.A | Pis_GEN_58_3_4_1/OPAF20776 | |||
| Oc9_C4 | V | OPAD021282/OPK181049 | |||
| Ascochyta blight resistance | Af1 | III | Vf6 × Vf136 (F2) | OPA111045/OPAB071026 | [90] |
| Af2 | II | OPE171272/OPJ18626 | |||
| Af3 | III | 29H × Vf136 (F2) | OPD161732/OPG041131 | [89] | |
| Af4 | unknown | OPJ18655/OPG111118 | |||
| Af1 (DSL) b | III.A | Vf6 × Vf136 (RILs) | OPAC061023 | [93] | |
| Af1 (DSS) c | III.A | OPF08710 | |||
| Af2 (DSL) | II.A | OPAG05737/Mer04790 | |||
| Af2 (DSS) | II.A | OPD12425/OPE171326 | |||
| Af1 (DSL) | III.A | Vf6 × Vf136 (RILs) | OPZ82 | [92] | |
| Af1 (DSS) | III.A | OPZ82 | |||
| Af2 (DSL) | II.A | OPAG05737/Mer04790 | |||
| Af2 (DSS) | II.A | OPD12425/OPE171326 | |||
| Frost tolerance | U_AUSPC-1 | unknown | Côte d’Or 1 × BPL 4628 (RILs) | U091499/B20803 | [71] |
| U_AUSPC-2 | unknown | F15476/I10661 | |||
| U_AUSPC-3 | unknown | O18715/O18737 | |||
| H_AUSPC-1 | unknown | B13560 | |||
| H_AUSPC-2 | unknown | I10661/I06425 | |||
| Determinate growth habit | Vf_TFL1 | V | CAPS-TFL1 (Hin1II) * | [10] | |
| Ti-dCAPS * | [11] | ||||
| Zero tannins | zt-1 | unknown | Vf6 × zt-1 (F2) | SCC5551/SCG111171 | [13] |
| zt-2 | unknown | Vf6 × zt-2 (F2) | SCAD16-B565/SCAD16-H385 | [14] | |
| Low vicine-convicine | vc- | unknown | Vf6 × vc- (F2) | SCH01620/SCAB12850 | [12] |
4.1.2. Markers for Rust Resistance
4.1.3. Markers for Broomrape Resistance
4.1.4. Markers for Anti-Nutritional Characters
4.1.5. Markers for Determinate Growth Habit
4.2. Use of Genomic Technologies
| Species | Genome Size | Ploidy Level |
|---|---|---|
| Cicer arietinum (chickpea) | 740 Mbp | 2n = 16 |
| Lens culinaris (lentil) | 4 Gbp | 2n = 14 |
| Pisum sativum (field pea) | 5 Gbp | 2n = 14 |
| Vicia faba (faba bean) | 13 Gbp | 2n = 12 |
4.2.1. Comparative Genomics
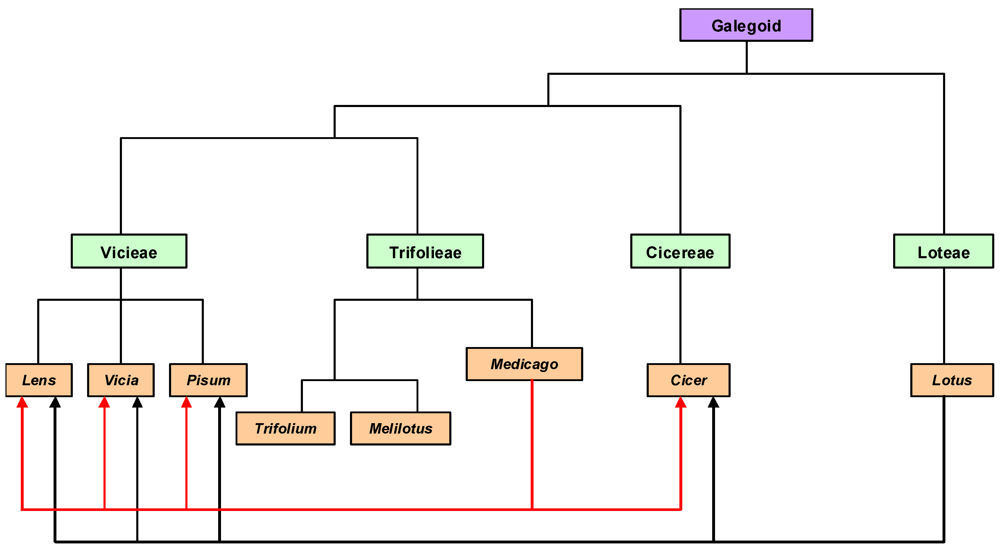
4.2.2. Functional Genomics
4.3. Genetic Transformation
4.3.1. In vitro Regeneration
4.3.2. Genetic Transformation
5. The Impact of Molecular Technologies on Faba Bean Breeding
5.1. New Approaches for More Efficient Targeted Marker Development
5.2. New Perspectives for Effective MAS
5.3. Identification and Characterisation of Candidate Genes
6. Future Perspectives
Acknowledgments
References and Notes
- Crépona, K.; Marget, P.; Peyronnet, C.; Carrouéea, B.; Arese, P.; Duc, G. Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crop. Res. 2010, 115, 329–339. [Google Scholar] [CrossRef]
- FAOSTAT. 2010 Production—Crops. Available online: http://faostat.fao.org (accessed on 2 May 2012).
- Roman, B.; Torres, A.M.; Rubiales, D.; Cubero, J.I.; Satovic, Z. Mapping of quantitative trait loci controlling broomrape (Orobanche crenata Forsk.) resistance in faba bean (Vicia faba L.). Genome 2002, 45, 1057–1063. [Google Scholar] [CrossRef]
- Torres, A.M.; Roman, B.; Avila, C.M.; Satovic, Z.; Rubiales, D.; Sillero, J.C.; Cubero, J.I.; Moreno, M.T. Faba bean breeding for resistance against biotic stresses: Towards application of marker technology. Euphytica 2006, 147, 67–80. [Google Scholar] [CrossRef]
- Ellwood, S.R.; Phan, H.T.; Jordan, M.; Hane, J.; Torres, A.M.; Avila, C.M.; Cruz-Izquierdo, S.; Oliver, R.P. Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 2008, 9, 380. [Google Scholar]
- van de Ven, W.T.G.; Waugh, R.; Duncan, N.; Ramsay, G.; Dow, N.; Powell, W. Development of a genetic linkage map in Vicia faba using molecular and biochemical techniques. Asp. Appl. Biol. 1991, 27, 49–54. [Google Scholar]
- Ramsay, G.; van de Ven, W.; Waugh, R.; Griffiths, D.W.; Powell, W. AEP (l’Association Européene de Recherche sur les Protéagineuse). Mapping quantitative trait loci in Faba beans. In Proceedings of the 2nd European Conference on Grain Legumes, Copenhagen, Denmark; Copenhagen, Denmark: 1995; pp. 444–445.
- Satovic, Z.; Torres, A.M.; Cubero, J.I. Genetic mapping of new morphological, isozyme and RAPD markers in Vicia faba L. using trisomics. Theor. Appl. Genet. 1996, 93, 1130–1138. [Google Scholar] [CrossRef]
- Torres, A.M.; Avila, C.M.; Gutierrez, N.; Palomino, C.; Moreno, M.T.; Cubero, J.I. Marker-assisted selection in faba bean (Vicia faba L.). Field Crop. Res. 2010, 115, 243–252. [Google Scholar] [CrossRef]
- Avila, C.M.; Nadal, S.; Moreno, M.T.; Torres, A.M. Development of a simple PCR-based marker for the determination of growth habit in Vicia faba L. using a candidate gene approach. Mol. Breed. 2006, 17, 185–190. [Google Scholar] [CrossRef]
- Avila, C.M.; Atienza, S.G.; Moreno, M.T.; Torres, A.M. Development of a new diagnostic marker for growth habit selection in faba bean (Vicia faba L.) breeding. Theor. Appl. Genet. 2007, 115, 1075–1082. [Google Scholar]
- Gutierrez, N.; Avila, C.M.; Duc, G.; Marget, P.; Suso, M.J.; Moreno, M.T.; Torres, A.M. Markers to assist selection for low vicine and convicine contents in faba bean (Vicia faba L.). Theor. Appl. Genet. 2006, 114, 59–66. [Google Scholar]
- Gutierrez, N.; Avila, C.; Rodriguez-Suarez, C.; Moreno, M.; Torres, A. Development of SCAR markers linked to a gene controlling absence of tannins in faba bean. Mol. Breed. 2007, 19, 305–314. [Google Scholar]
- Gutierrez, N.; Avila, C.M.; Moreno, M.T.; Torres, A.M. Development of SCAR markers linked to zt-2, one of the genes controlling absence of tannins in faba bean. Aust. J. Agric. Res. 2008, 59, 62–68. [Google Scholar]
- Tanno, K.; Willcox, G. How fast was wild wheat domesticated? Science 2006, 311, 1886. [Google Scholar]
- Cubero, J.I. On the evolution of Vicia faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar]
- Paull, J.G.; Kimber, R.; van Leur, J. Faba bean breeding and production in Australia. Grain Legum. 2011, 56, 15–16. [Google Scholar]
- Duc, G.; Bao, S.; Baum, M.; Redden, R.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crops Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Bond, D.A. Recent developments in breeding field beans (Vicia faba L.). Plant Breed. 1987, 99, 1–26. [Google Scholar] [CrossRef]
- Briggs, F.N.; Knowles, P.F. Introduction to Plant Breeding; Reinhold Publishing Corporation: New York, NY, USA, 1967; p. 426. [Google Scholar]
- Link, W.; Schill, B.; Kittlitz, E.V. Breeding for wide adaptation in faba bean. Euphytica 1996, 92, 185–190. [Google Scholar] [CrossRef]
- Bond, D.A. Prospects for commercialisation of F1 hybrid field beans Vicia faba L. Euphytica 1989, 41, 81–86. [Google Scholar] [CrossRef]
- Link, W.; Ederer, W.; Gumber, R.K.; Melchinger, A.E. Detection and characterization of two new CMS systems in faba bean (Vicia faba). Plant Breed. 1997, 116, 158–162. [Google Scholar]
- Hanounik, S.B.; Robertson, L.D. Resistance in Vicia faba germplasm to blight caused by Ascochyta fabae. Plant Dis. 1989, 73, 202–205. [Google Scholar] [CrossRef]
- Kittlitz, E.V.; Ibrahim, K.I.M.; Ruckenbauer, P.; Robertson, L.D. Analysis and use of inter-pool crosses (Mediterranean × Central European) in faba beans (Vicia faba L.). Plant Breed. 1993, 110, 307–314. [Google Scholar]
- Annicchiarico, P.; Iannucci, A. Breeding strategy for faba bean in Southern Europe based on cultivar responses across climatically contrasting environments. Crop Sci. 2008, 48, 983–991. [Google Scholar] [CrossRef]
- Bao, S. Yunnan Academy of Agricultural Sciences: Kunming, China. Personal communication, 2004.
- Frauen, M.; Sass, O. Inheritance and performance of the stiff-strawed mutant in Vicia faba L. In Proceedings of XII EUCARPIA Congress, Göttingen, Germany, 1989; 15, pp. 13–18.
- Nadal, S.; Moreno, M.T.; Cubero, J.I. Registration of “Retaca” faba bean. Crop Sci. 2004, 44, 1865. [Google Scholar] [CrossRef]
- Adisarwanto, T.; Knight, R. Effect of sowing date and plant density on yield and yield components in the faba bean. Aust. J. Agric. Res. 1997, 48, 1161–1168. [Google Scholar] [CrossRef]
- Loss, S.P.; Siddique, K.H.M.; Jettner, R.; Martin, L.D. Responses of faba bean (Vicia faba L.) to sowing rate in south-western Australia. I. Seed yield and economic optimum plant density. Aust. J. Agric. Res. 1998, 49, 989–997. [Google Scholar] [CrossRef]
- Sillero, J.C.; Villegas-Fernández, A.M.; Thomas, J.; Rojas-Molina, M.M.; Emeran, A.A.; Fernández-Aparicio, M.; Rubiales, D. Faba bean breeding for disease resistance. Field Crops Res. 2010, 115, 297–307. [Google Scholar] [CrossRef]
- Kharbanda, P.D.; Bernier, C.C. Cultural and pathogenic variability among isolates of Ascochyta fabae. Can. J. Plant Pathol. 1980, 2, 139–142. [Google Scholar] [CrossRef]
- Sillero, J.C.; Morenoa, M.T.; Rubialesb, D. Characterization of new sources of resistance to Uromyces viciae-fabae in a germplasm collection of Vicia faba. Plant Pathol. 2000, 49, 389–395. [Google Scholar] [CrossRef]
- Hanounik, S.B.; Robertson, L.D. Resistance in Vicia faba plasm to blight caused by Ascochyta fabae. Plant Disease 1989, 73, 202–205. [Google Scholar] [CrossRef]
- Kimber, R.B.E.; Davidson, J.A.; Paull, J.G. Using genetic diversity within faba bean germplasm to develop resistance to ascochyta blight. In Proceedings of the1st International Ascochyta Workshop on Grain Legumes, Le Trodet, France, 2–6 July 2006.
- Bond, D.A.; Jellis, G.J.; Rowland, G.G.; Le Guen, J.; Robertson, L.D.; Khalil, S.A.; Li-Juan, L. Present status and future strategy in breeding faba beans (Vicia faba L.) for resistance to biotic and abiotic stresses. Euphytica 1994, 73, 151–166. [Google Scholar] [CrossRef]
- Hanounik, S.B.; Robertson, L.D. New sources of resistance in Vicia faba to chocolate spot caused by Botrytis fabae. Plant Dis. 1988, 72, 696–698. [Google Scholar] [CrossRef]
- Bouhassan, A.; Sadiki, M.; Tivoli, B. Evaluation of a collection of faba bean (Vicia faba L.) genotypes originating from the Maghreb for resistance to chocolate spot (Botrytis fabae) by assessment in the field and laboratory. Euphytica 2004, 135, 55–62. [Google Scholar] [CrossRef]
- Paull, J.G. University of Adelaide: Adelaide, Australia. Personal observation, 2012.
- Rashid, K.Y.; Bernier, C.C.; Conner, R.L. Evaluation of fava bean for resistance to Ascochyta fabae and development of host differentials for race identification. Plant Dis. 1991, 75, 852–855. [Google Scholar] [CrossRef]
- Kohpina, S.; Knight, R.; Stoddard, F.L. Variability of Ascochyta fabae in South Australia. Aust. J. Agric. Res. 1999, 50, 1475–1481. [Google Scholar] [CrossRef]
- Hanounik, S.B.; Maliha, N. Horizontal and vertical resistance in Vicia faba to chocolate spot caused by Botrytis fabae. Plant Dis. 1986, 70, 770–773. [Google Scholar] [CrossRef]
- Rashid, K.Y.; Bernier, C.C. Evaluation of resistance in Vicia faba to two isolates of the rust fungus Uromyces viciae-fabae from Manitoba. Plant Dis. 1984, 68, 16–18. [Google Scholar]
- Villegas-Fernández, A.M.; Sillero, J.C.; Emeran, A.A.; Flores, F.; Rubiales, D. Multiple-disease resistance in Vicia faba: multi-environment field testing for identification of combined resistance to rust and chocolate spot. Field Crops Res. 2011, 124, 59–65. [Google Scholar] [CrossRef]
- Rubiales, D.; Fernández-Aparicio, M. Innovations in parasitic weeds management in legume crops. A review. Agron. Sustain. Dev. 2012, 32, 433–449. [Google Scholar] [CrossRef]
- Gressel, J.; Hanafi, A.; Head, G.; Marasas, W.; Obilana, A.B.; Ochanda, J.; Souissi, T.; Tzotzos, G. Major heretofore intractable biotic constraints to African food security that may be amenable to novel biotechnological solutions. Crop Prot. 2004, 23, 661–689. [Google Scholar] [CrossRef]
- Pérez-de-Luque, A.; Eizenberg, H.; Grenz, J.H.; Sillero, J.C.; Ãvila, C.; Sauerborn, J.; Rubiales, D. Broomrape management in faba bean. Field Crops Res. 2010, 115, 319–328. [Google Scholar] [CrossRef]
- Joel, D.M. The long-term approach to parasitic weeds control: manipulation of specific developmental mechanisms of the parasite. Crop Protect. 2000, 19, 753–758. [Google Scholar] [CrossRef]
- Mauromicale, G.; Restuccia, G.; Marchese, M. Soil solarization, a nonchemical technique for controlling Orobanche crenata and improving yield of faba bean. Agronomie 2001, 21, 757–765. [Google Scholar] [CrossRef]
- Rubiales, D. Parasitic plants, wild relatives and the nature of resistance. New Phytol. 2003, 160, 459–461. [Google Scholar] [CrossRef]
- Stoddard, F.L.; Nicholas, A.H.; Rubiales, D.; Thomas, J.; Villegas-Fernández, A.M. Integrated pest management in faba bean. Field Crops Res. 2010, 115, 308–318. [Google Scholar] [CrossRef]
- Cubero, J.I. Breeding for resistance to Orobanche species: A review. In Proceedings of the International Workshop on Orobanche Research, Obermarchtal, Germany, 19–22 August 1989; Wegmann, K., Musselman, L.J., Eds.; Eberhard-Karls-Universität: Tübingen, Germany, 1991; pp. 257–277. [Google Scholar]
- Sillero, J.C.; Rubiales, D.; Cubero, J.I. Risks of Orobanche screenings based only on final number of emerged shoots per plant. In Advances in Parasitic Plant Research; Moreno, M.T., Cubero, J.I., Berner, D., Joel, D., Musselman, L.J., Parker, C., Eds.; Junta de Andalucìa: Córdoba, Spain, 1996; pp. 652–657. [Google Scholar]
- Cubero, J.I.; Moreno, M.T. Studies on resistance to Orobanche crenata in Vicia faba. In Resistance to Broomrape—The State of the Art; Cubero, J.I., Moreno, M.T., Rubiales, D., Sillero, J.C., Eds.; DGIFA: Junta de Andalucía, Sevilla, Spain, 1999; pp. 9–15. [Google Scholar]
- Muehlbauer, F.J.; Kaiser, W.J.; Simon, C.J. Potential for wild species in cool season food legume breeding. Euphytica 1993, 73, 109–114. [Google Scholar]
- Khan, H.R.; Paull, J.G.; Siddique, K.H.M.; Stoddard, F.L. Faba bean breeding for drought-affected environments: A physiological and agronomic perspective. Field Crops Res. 2010, 115, 279–286. [Google Scholar] [CrossRef]
- French, R.J. The risk of vegetative water deficit in early-sown faba bean (Vicia faba L.) and its implications for crop productivity in a Mediterranean-type environment. Crop Pasture Sci. 2010, 61, 566–577. [Google Scholar] [CrossRef]
- Oweis, T.; Hachum, A.; Pala, M. Faba bean productivity under rainfed and supplemental irrigation in northern Syria. Agr. Water Manag. 2005, 73, 57–72. [Google Scholar] [CrossRef]
- Solaiman, Z.; Colmer, T.D.; Loss, S.P.; Thomson, B.D.; Siddique, K.H.M. Growth responses of cool-season grain legumes to transient waterlogging. Aust. J. Agric. Res. 2007, 58, 406–412. [Google Scholar] [CrossRef]
- Loss, S.P.; Siddique, K.H.M.; Tennant, D. Adaptation of faba bean (Vicia faba L.) to dryland Mediterranean-type environments. III. Water use and water-use efficiency. Field Crop. Res. 1997, 54, 153–162. [Google Scholar] [CrossRef]
- Turpin, J.E.; Robertson, M.J.; Hillcoat, N.S.; Herridge, D.F. Faba bean (Vicia faba) in Australia's northern grains belt: Canopy development, biomass, and nitrogen accumulation and partitioning. Aust. J. Agric. Res. 2002, 53, 227–237. [Google Scholar] [CrossRef]
- Khan, H.R.; Paull, J.G.; Siddique, K.H.M.; Stoddard, F.L. Faba bean breeding for drought-affected environments: A physiological and agronomic perspective. Field Crops Res. 2009, 115, 279–286. [Google Scholar]
- Khan, H.R.; Link, W.; Hocking, T.J.H.; Stoddard, F.L. Evaluation of physiological traits for improving drought tolerance in faba bean (Vicia faba L.). Plant Soil 2007, 292, 205–217. [Google Scholar] [CrossRef]
- Stelling, D. Heterosis and hybrid performance in topless faba beans (Vicia faba L.). Euphytica 1997, 97, 73–79. [Google Scholar]
- Boddi, M.; Enneking, D.; Materne, M.; Paull, J.; Noy, D. Genetic variability in faba beans (Vicia faba) in response to NaCl. Contemporary crop improvement—A tropical view. In Proceedings of the14th Australasian Plant Breeding (APB) Conference and 11th Society for the Advancement of Breeding Researches in Asia and Oceania (SABRAO) Conference, Cairns, Australia, 10–14 August 2009.
- Tavakkoli, E.; Paull, J.; Rengasamy, P.; McDonald, G.K. Comparing genotypic variation in faba bean (Vicia faba L.) in response to salinity in hydroponic and field experiments. Field Crop. Res. 2012, 127, 99–108. [Google Scholar] [CrossRef]
- Rathjen, A.H. The Response of Grain Legumes to Boron.
- Hall, A.E. Breeding for heat tolerance. Plant Breed. Rev. 1992, 10, 129–168. [Google Scholar]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Env. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Arbaoui, M.; Balko, C.; Link, W. Study of faba bean (Vicia faba L.) winter-hardiness and development of screening methods. Field Crop. Res. 2008, 106, 60–67. [Google Scholar] [CrossRef]
- Inci, N.E.; Toker, C. Screening and selection of faba beans (Vicia faba L.) for cold tolerance and comparison to wild relatives. Genet. Resour. Crop Evol. 2011, 58, 1169–1175. [Google Scholar] [CrossRef]
- Nassar-Abbas, S.M. Investigation of Environmental Staining and Storage on Discolouration and Cooking Quality in Faba Bean (Vicia faba L.).
- Larralde, J.; Martinez, J.A. Nutritional value of faba bean: Effects on nutrient utilization, protein turnover and immunity. Options Méditerr. 1991, 10, 111–117. [Google Scholar]
- El-Sherbeeny, M.; Robertson, L.D. Protein content variation in a pure line faba bean (Vicia faba) collection. J. Sci. Food Agric. 2006, 58, 193–196. [Google Scholar] [CrossRef]
- Link, W. Methods and objectives in fababean breeding. In Proceedings of the International Workshop on Faba Bean Breeding and Agronomy, Córdoba, Spain, 25–27 October 2006; Junta de Andalucia: Córdoba, Spain, 2006; pp. 35–40. [Google Scholar]
- Kuman, R.; Singh, M. Tannins: Their adverse role in ruman nutrition. J. Agric. Food Chem. 1984, 32, 447. [Google Scholar] [CrossRef]
- Bartolomé, B.; Quesada, C.; Gómez-Cordibés, C.; Hernandez, T.; Estrella, I. New contributions to the inhibition study of R-amylase and trypsin by phenolic compounds. In Bioactive Substances in Food of Plant Origin; Kozlowska, H., Fornal, J., Zdunczyk, Eds.; Polish Academy of Sciences: Olstyn, Poland, 1994; Volume 1, pp. 233–238. [Google Scholar]
- Nelson, L.D.; Cox, M. Glycolysis, gluconeogenesis, and the pentose phosphate pathway. In Principles of Biochemistry; Freeman: New York, NY, USA, 2005; p. 551. [Google Scholar]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. Soc. Biol. Sci. 2008, 363, 557–572. [Google Scholar]
- Johnson, G.R.; McCuddin, Z.P. Maize and the biotech industry. In Handbook of Maize: Its Biology; Bennetzen, J.L., Hake, S.C., Eds.; Springer: Berlin, Germany, 2009; pp. 115–140. [Google Scholar]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Torres, A.M.; Weeden, N.F.; Martin, A. Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theor. Appl. Genet. 1993, 85, 937–945. [Google Scholar]
- Pozarkova, D.; Koblizkova, A.; Román, B.; Torres, A.M.; Lucretti, S.; Lysak, M.; Dolezel, J.; Macas, J. Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biol. Plant. 2002, 45, 337–345. [Google Scholar] [CrossRef]
- Alghamdi, S.; Migdadi, H.; Ammar, M.; Paull, J.; Siddique, K.H.M. Faba bean genomics: Current status and future prospects. Euphytica 2012. [Google Scholar]
- Avila, C.M.; Sillero, J.C.; Rubiales, D.; Moreno, M.T.; Torres, A.M. Identification of RAPD markers linked to the Uvf-1 gene conferring hypersensitive resistance against rust (Uromyces viciae-fabae) in Vicia faba L. Theor. Appl. Genet. 2003, 107, 353–358. [Google Scholar] [CrossRef]
- Roman, B.; Alfaro, C.; Torres, A.M.; Moreno, M.T.; Satovic, Z.; Pujadas, A.; Rubiales, D. Genetic relationships among Orobanche species as revealed by RAPD analysis. Ann. Bot. 2003, 91, 637–642. [Google Scholar] [CrossRef]
- Avila, C.M.; Satovic, Z.; Sillero, J.C.; Rubiales, D.; Moreno, M.T.; Torres, A.M. Isolate and organ-specific QTLs for ascochyta blight resistance in faba bean (Vicia faba L.). Theor. Appl. Genet. 2004, 108, 1071–1078. [Google Scholar] [CrossRef]
- Roman, B.; Satovic, Z.; Avila, C.M.; Rubiales, D.; Moreno, M.T.; Torres, A.M. Locating genes associated with Ascochyta fabae resistance in Vicia faba. Aust. J. Agric. Res. 2003, 54, 85–90. [Google Scholar]
- Diaz-Ruiz, R.; Torres, A.M.; Satovic, Z.; Gutierrez, M.V.; Cubero, J.I.; Roman, B. Validation of QTLs for Orobanche crenata resistance in faba bean (Vicia faba L.) across environments and generations. Theor. Appl. Genet. 2010, 120, 909–919. [Google Scholar] [CrossRef]
- Satovic, Z.; Avila, C.; Palomino, C.; Vitale, S.; Gutierrez, N.; Cruz-Izquierdo, S.; Gutierrez, M.V.; Ruiz, M.D.; Ocaña, S.; Torres, A.M. Towards a unified and functional consensus linkage map in faba bean (Vicia faba L.). Candidate Gene Identification and Breeding Applications, 2012.
- Díaz-Ruiz, R.; Satovic, Z.; Avila, C.M.; Alfaro, C.M.; Gutierrez, M.V.; Torres, A.M.; Román, B. Confirmation of QTLs controlling Ascochyta fabae resistance in different generations of faba bean (Vicia faba L.). Crop Pasture Sci. 2009, 60, 353–361. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef]
- Sillero, J.C.; Fondevilla, S.; Davidson, J.; Vaz Patto, M.C.; Warkentin, T.; Thomas, J.; Rubiales, D. Screening techniques and sources of resistance to rusts and mildews in grain legumes. Euphytica 2006, 147, 255–272. [Google Scholar] [CrossRef]
- Duvick, D.N. Plant breeding, an evolutionary concept. Crop Sci. 1996, 36, 539–548. [Google Scholar] [CrossRef]
- Roman, B.; Satovic, Z.; Rubiales, D.; Torres, A.M.; Cubero, J.I.; Katzir, N.; Joel, D.M. Variation among and within populations of the parasitic weed Orobanche crenata from Spain and Israel revealed by Inter Simple Sequence Repeat markers. Phytopathology 2002, 92, 1262–1266. [Google Scholar] [CrossRef]
- Nassib, A.M.; Ibrahim, A.A.; Khalil, S.A. Breeding for resistance to Orobanche. In Faba Bean Improvement; Hawtin, G., Webb, C., Eds.; Martinus Nijhoff: Hague, The Netherlands, 1982; pp. 199–206. [Google Scholar]
- Cubero, J.I.; Moreno, M.T.; Hernández, L. A faba bean (Vicia faba L.) cultivar resistant to broomrape (Orobanche crenata Forsk.). In Proceedings of the First Europe Conference on Grain Legumes; Angers, F., Ed.; Association Europeenne des Protéagineux: Paris, France, 1992; pp. 41–42. [Google Scholar]
- Austin, D.F.; Lee, M. Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor. Appl. Genet. 1996, 92, 817–826. [Google Scholar] [CrossRef]
- Duc, G.; Sixdenier, G.; Lila, M.; Furstoss, V. Search of Genetic Variability for Vicine and Convicine Content in Vicia faba L. A first report of a gene which codes for nearly zero-vicine and zero-convicine contents. In Recent Advances of Research in Antinutritional Factors in Legume Seeds; Huisman, J., van der Poel, A.F.B., Liener, I.E., Eds.; Pudoc: Wageningen, The Netherlands, 1989; pp. 305–313. [Google Scholar]
- Gutierrez, N.; Avila, C.M.; Duc, G.; Marget, P.; Suso, M.J.; Moreno, M.T.; Torres, A.M. CAPs markers to assist selection for low vicine and convicine contents in faba bean (Vicia faba L.). Theor. Appl. Genet. 2006, 114, 59–66. [Google Scholar] [CrossRef]
- Cabrera, A.; Martin, A. Variation in tannin content in Vicia faba L. J. Agric. Sci. 1986, 106, 377–382. [Google Scholar] [CrossRef]
- Duc, G.; Marget, P.; Esnault, R.; Le Guen, J.; Bastianelli, D. Genetic variability for feeding value of faba bean seeds (Vicia faba): Comparative chemical composition of isogenics involving zero-tannin and zero-vicine genes. J. Agric. Sci. 1999, 133, 185–196. [Google Scholar] [CrossRef]
- Varshney, R.K.; Close, T.J.; Singh, N.K.; Hoisington, D.A.; Cook, D.R. Orphan legume crops enter the genomics era! Curr. Opin. Plant. Biol. 2009, 12, 202–210. [Google Scholar] [CrossRef]
- Kaur, S.; Cogan, N.O.; Pembleton, L.W.; Shinozuka, M.; Savin, K.W.; Materne, M.; Forster, J.W. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics 2011, 12, 265. [Google Scholar]
- Kaur, S.; Pembleton, L.; Cogan, N.; Savin, K.; Leonforte, T.; Paull, J.; Materne, M.; Forster, J. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 2012, 13, 104. [Google Scholar]
- Sato, S.; Nakamura, Y.; Asamizu, E.; Isobe, S.; Tabata, S. Genome sequencing and genome resources in model legumes. Plant Physiol. 2007, 144, 588–593. [Google Scholar] [CrossRef]
- Sandal, N.; Petersen, T.R.; Murray, J.; Umehara, Y.; Karas, B.; Yano, K.; Kumagai, H.; Yoshikawa, M.; Saito, K.; Hayashi, M.; et al. Genetics of symbiosis in Lotus japonicus: Recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 2006, 19, 80–91. [Google Scholar] [CrossRef]
- Choi, H.-K.; Mun, J.-H.; Kim, D.-J.; Zhu, H.; Baek, J.-M.; Mudge, J.; Roe, B.; Ellis, N.; Doyle, J.; Kiss, G.B.; et al. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA 2004, 101, 15289–15294. [Google Scholar]
- Wang, X.; Sato, S.; Tabata, S.; Kawasaki, S. A high-density linkage map of Lotus japonicus based on AFLP and SSR markers. DNA Res. 2008, 15, 323–332. [Google Scholar] [CrossRef]
- Gallardo, K.; Firnhaber, C.; Zuber, H.; Hericher, D.; Belghazi, M.; Henry, C.; Kuster, H.; Thompson, R. A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol. Cell Proteomics 2007, 6, 2165–2179. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Lippold, F.; Redestig, H.; Hannah, M.A.; Erban, A.; Kramer, U.; Kopka, J.; Udvardi, M.K. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 2008, 53, 973–987. [Google Scholar]
- Watson, B.S.; Asirvatham, V.S.; Wang, L.; Sumner, L.W. Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol. 2003, 131, 1104–1123. [Google Scholar] [CrossRef]
- Gonzales, M.D.; Archuleta, E.; Farmer, A.; Gajendran, K.; Grant, D.; Shoemaker, R.; Beavis, W.D.; Waugh, M.E. The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res. 2005, 33, D660–D665. [Google Scholar]
- Rispail, N.; Péter, K.; Kiss, G.B.; Ellis, T.H.N.; Gallardo, K.; Thompson, R.D.; Prats, E.; Larrainzar, E.; Ladrera, R.; González, E.M.; et al. Model legumes contribute to faba bean breeding. Field Crops Res. 2010, 115, 253–269. [Google Scholar] [CrossRef]
- Kaló, P.; Endre, G.; Zimányi, L.; Csanádi, G.; Kiss, G.B. Construction of an improved linkage map of diploid alfalfa (Medicago sativa). Theor. Appl. Genet. 2000, 100, 641–657. [Google Scholar] [CrossRef]
- Cannon, S.B.; Sterck, L.; Rombauts, S.; Sato, S.; Cheung, F.; Gouzy, J.; Wang, X.; Mudge, J.; Vasdewani, J.; Schiex, T.; et al. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 14959–14964. [Google Scholar]
- Phan, H.T.; Ellwood, S.R.; Hane, J.K.; Ford, R.; Materne, M.; Oliver, R.P. Extensive macrosynteny between Medicago truncatula and Lens culinaris ssp. culinaris. Theor. Appl. Genet. 2007, 114, 549–558. [Google Scholar] [CrossRef]
- Roman, B.; Satovic, Z.; Pozarkova, D.; Macas, J.; Dolezel, J.; Cubero, J.I.; Torres, A.M. Development of a composite map in Vicia faba, breeding applications and future prospects. Theor. Appl. Genet. 2004, 108, 1079–1088. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar]
- Chen, Y.-L.; Huang, R.; Xiao, Y.-M.; Lu, P.; Chen, J.; Wang, X.-C. Extracellular calmodulin-induced stomatal closure is mediated by heterotrimeric G protein and H2O2. Plant Physiol. 2004, 136, 4096–4103. [Google Scholar] [CrossRef]
- Gao, X.Q.; Li, C.G.; Wei, P.C.; Zhang, X.Y.; Chen, J.; Wang, X.C. The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol. 2005, 139, 1207–1216. [Google Scholar]
- Perlick, A.M.; Frühling, M.; Schröder, G.; Frosch, S.C.; Pühler, A. The broad bean gene VfNOD32 encodes a nodulin with sequence similarities to chitinases that is homologous to (alpha/beta) 8-barrel-type seed proteins. Plant Physiol. 1996, 110, 147–154. [Google Scholar]
- Hanstein, S.M.; Felle, H.H. CO2-triggered chloride release from guard cells in intact fava bean leaves. Kinetics of the onset of stomatal closure. Plant Physiol. 2002, 130, 940–950. [Google Scholar] [CrossRef]
- Li, J.; Assmann, S.M. An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 1996, 8, 2359–2368. [Google Scholar]
- Iwai, S.; Shimomura, N.; Nakashima, A.; Etoh, T. New fava bean guard cell signaling mutant impaired in ABA-induced stomatal closure. Plant Cell Physiol. 2003, 44, 909–913. [Google Scholar] [CrossRef]
- Miranda, M.; Borisjuk, L.; Tewes, A.; Heim, U.; Sauer, N.; Wobus, U.; Weber, H. Amino acid permeases in developing seeds of Vicia faba L.: Expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J. 2001, 28, 61–71. [Google Scholar] [CrossRef]
- Horstmann, C.; Schlesier, B.; Otto, A.; Kostka, S.; Müntz, K. Polymorphism of legumin subunits from field bean (Vicia faba L. var. minor) and its relation to the corresponding multigene family. Theor. Appl. Genet. 1993, 86, 867–874. [Google Scholar] [CrossRef]
- Weschke, W.; Bassüner, R.; Van Hai, N.; Czihal, A.; Baümlein, H.; Wobus, U. The structure of a Vicia faba vicilin gene. Biochem. Physiol. Pflanzen. 1988, 183, 233–242. [Google Scholar]
- Tucci, M.; Capparelli, R.; Costa, A.; Rao, R. Molecular heterogeneity and genetics of Vicia faba seed storage proteins. Theor. Appl. Genet. 1991, 81, 50–58. [Google Scholar]
- Knaak, C.; Roskothen, P.; Roebbelen, G. Symbiotic efficiency of Vicia faba genotypes after field inoculation with different strains of Rhizobium leguminosarum preselected in greenhouse tests. J. Plant Physiol. 1993, 141, 49–53. [Google Scholar] [CrossRef]
- Hohnjec, N.; Küster, H.; Albus, U.; Frosch, S.C.; Becker, J.D.; Pühler, A.; Perlick, A.M.; Frühling, M. The broad bean nodulin VfENOD18 is a member of a novel family of plant proteins with homologies to the bacterial MJ0577 superfamily. Mol. Gen. Genet. 2000, 264, 241–250. [Google Scholar] [CrossRef]
- Perlick, A.M.; Pühler, A. A survey of transcripts expressed specifically in root nodules of broadbean (Vicia faba L.). Plant Mol. Biol. 1993, 22, 957–970. [Google Scholar] [CrossRef]
- Küster, H.; Frühling, M.; Perlick, A.M.; Pühler, A. The sucrose synthase gene is predominantly expressed in the root nodule tissue of Vicia faba. Mol Plant Microbe Interact. 1993, 6, 507–514. [Google Scholar] [CrossRef]
- Schroder, G.; Fruhling, M.; Puhler, A.; Perlick, A.M. The temporal and spatial transcription pattern in root nodules of Vicia faba nodulin genes encoding glycine-rich proteins. Plant Mol. Biol. 1997, 33, 113–123. [Google Scholar] [CrossRef]
- Fehlberg, V.; Vieweg, M.F.; Dohmann, E.M.; Hohnjec, N.; Puhler, A.; Perlick, A.M.; Kuster, H. The promoter of the leghaemoglobin gene VfLb29: Functional analysis and identification of modules necessary for its activation in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots. J. Exp. Bot. 2005, 56, 799–806. [Google Scholar] [CrossRef]
- Frühling, M.; Roussel, H.; Gianinazzi-Pearson, V.; Pühler, A.; Perlick, A.M. The Vicia faba leghemoglobin gene VfLb29 is induced in root nodules and in roots colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. Mol. Plant. Microbe Interact. 1997, 10, 124–131. [Google Scholar]
- Vieweg, M.F.; Frühling, M.; Quandt, H.J.; Heim, U.; Bäumlein, H.; Pühler, A.; Küster, H.; Andreas, M.P. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol. Plant Microbe Interact. 2004, 17, 62–69. [Google Scholar] [CrossRef]
- Pickersgill, B.; Jones, J.K.; Ramsay, G.; Stewart, H. Problems and prospects of wild crossing in the genus Vicia for the improvement of faba bean. In Proceedings of the International Worshop on Faba beansKabuli Chickpeas and Lentil in the 1980s, Aleppo, Syria, 16–20 May 1983; Saxena, M.C., Varma, S., Eds.; ICARDA: Aleppo, Syria, 1983. [Google Scholar]
- Ramsay, G.; Pickersgill, B.; Jones, J.K.; Hammond, L.; Stewart, M.H. Barriers to interspecific hybridization between V. faba and other species of section Faba. In Vicia faba: Agronomy, Physiology and Breeding; Hebblethwaite, P.D., Dawkins, T.C.K., Heath, M.C., Lockwood, G., Eds.; Martinus Nijhoff: Hague, The Netherlands, 1984; pp. 201–208. [Google Scholar]
- Ladizinsky, G.; Pickersgill, B.; Yamamoto, K. Exploitation of wild relatives of the food legumes. In World Crops: Cool Season Food Legumes; Summerfield, R.J., Ed.; Kluwer Academic Press: Dordrecht, The Netherlands, 1988; p. 967. [Google Scholar]
- Venketeswaran, S. Tissue culture studies on Vicia faba L. Establishment of culture. Phytomorphology 1962, 12, 300–306. [Google Scholar]
- Grant, M.; Fuller, K.W. Tissue culture of root cells of Vicia faba. J. Exp. Bot. 1968, 19, 667–680. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Gildow, F.E. The initiation and maintenance of Vicia faba tissue cultures. Physiol. Plant. 1975, 34, 250–253. [Google Scholar] [CrossRef]
- Röper, W. Growth and cytology of callus and cell suspension cultures of Vicia faba L. Z. Pflanzenphysiol. 1979, 93, 245–257. [Google Scholar]
- Bieri, V.; Schmid, J.; Keller, E.R. Shoot tip culture in Vicia faba L. In Efficiency in Plant Breeding: Proceedings of the 10th Congress of the European Association for Research on Plant Breeding, Wageningen, Netherlands, 19–24 June 1983; Lange, W., Zeven, A.C., Hogenboom, N.F., Eds.; EUCARPIA: Wageningen, The Netherlands, 1984; p. 295. [Google Scholar]
- Selva, E.; Stouffs, M.; Briquet, M. In vitro propagation of Vicia faba L. by micro-cutting and multiple shoot induction. Plant Cell Tiss. Org. Cult. 1989, 18, 167–179. [Google Scholar] [CrossRef]
- Thynn, M.; Werner, D. Plantlet regeneration and somatic differentiation in faba bean (Vicia faba L.) from callus culture of various explants. Angewandte Botanik 1987, 61, 483–492. [Google Scholar]
- Binding, H.; Nehls, R. Regeneration of isolated protoplasts of Vicia faba L. Z. Pflanzenphysiol. 1978, 88, 327–332. [Google Scholar]
- Binding, H.; Nehls, R. Somatic cell hybridization of Vicia faba and Petunia hybrida. Mol. Gen. Genet. 1978, 164, 137–143. [Google Scholar] [CrossRef]
- Röper, W. Callus formation from protoplasts derived from cell suspension cultures of Vicia faba L. . Z. Pflanzenphysiol. 1981, 101, 75–78. [Google Scholar]
- Tegeder, M.; Gebhardt, D.; Schieder, O.; Pickardt, T. Thidiazuron-induced plant regeneration from protoplasts of Vicia faba cv. Mythos. Plant Cell Rep. 1995, 15, 164–169. [Google Scholar]
- Griga, M.; Kubalakova, M.; Tejklova, E. Somatic embryogenesis in Vicia faba L. Plant Cell Tiss. Org. Cult. 1987, 9, 167–171. [Google Scholar] [CrossRef]
- Pickardt, T.; Huancaruna Perales, E.; Schieder, O. Plant regeneration via somatic embryogenesis in Vicia narbonensis. Protoplasma 1989, 149, 5–10. [Google Scholar] [CrossRef]
- Pickardt, T.; Meixner, M.; Schade, V.; Schieder, O. Transformation of Vicia narbonensis via Agrobacterium tumefaciens-mediated gene transfer. Plant Cell Rep. 1991, 9, 535–538. [Google Scholar]
- Jelenic, S.; Mitrikeski, P.T.; Papes, D.; Jelaska, S. Agrobacterium-mediated transformation of broad bean Vicia faba L. Food Technol. Biotech. 2000, 38, 167–172. [Google Scholar]
- Siefkes-Boer, H.J.; Noonan, M.J.; Bullock, D.W.; Conner, A.J. Hairy root transformation system in large- seeded grain legumes. Israel J. Plant Sci. 1995, 43, 1–5. [Google Scholar]
- Saalbach, I.; Pickardt, T.; Machemehl, F.; Saalbach, G.; Schieder, O.; Müntz, K. A chimeric gene encoding the methionine-rich 2S albumin of Brazil nut (Bertholletia excelsa H.B.K.) is stably expressed and inherited in transgenic grain legumes. Mol. Gen. Genet. 1994, 242, 226–236. [Google Scholar] [CrossRef]
- Schiemann, J.; Eisenreich, G. Transformation of field bean Vicia faba L. cells expression of a chimeric gene in cultured hairy roots and root-derived callus. Biochem. Physiol. Pflanzen. 1989, 185, 135–140. [Google Scholar]
- Ramsay, G.; Kumar, A. Transformation of Vicia faba cotyledon and stem tissues by Agrobacterium rhizogenes—Infectivity and cytological studies. J. Exp. Bot. 1990, 41, 841–847. [Google Scholar] [CrossRef]
- Böttinger, P.; Steinmetz, A.; Schieder, O.; Pickardt, T. Agrobacterium-mediated transformation of Vicia faba. Mol. Breed. 2001, 8, 243–254. [Google Scholar] [CrossRef]
- Hanafy, M.; Pickardt, T.; Kiesecker, H.; Jacobsen, H.-J. Agrobacterium-mediated transformation of faba bean (Vicia faba L.) using embryo axes. Euphytica 2005, 142, 227–236. [Google Scholar] [CrossRef]
- Cubero, J.I.; Nadal, S. Faba bean (Vicia faba L.). In Genetic Resources, Chromosome Engineering, and Crop Improvement for Grain legumes; Singh, R.J., Jauhar, P., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2005; p. 163. [Google Scholar]
- Palomino, C.; Satovic, Z.; Cubero, J.I.; Torres, A.M. Identification and characterization of NBS-LRR class resistance gene analogs in faba bean (Vicia faba L.) and chickpea (Cicer arietinum L.). Genome 2006, 49, 1227–1237. [Google Scholar] [CrossRef]
- Palomino, C.; Fernandez-Romero, M.D.; Rubio, J.; Torres, A.M.; Moreno, M.T.; Millán, T. Integration of new CAPS and dCAPS-RGA markers into a composite chickpea genetic map and their association with disease resistance. Theor. Appl. Genet. 2008, 118, 671–682. [Google Scholar]
- Grain Legumes Integrated Project (GLIP). Available online: http://www.eugrainlegumes.org/ (accessed on 13 January 2012).
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Crouch, J.H.; Mackill, D.J.; Xu, Y.; Blair, M.W.; Ragot, M.; Upadhyaya, H.D.; Ortiz, R. The molecularization of public sector crop breeding: Progress, problems, and prospects. Adv. Agron. 2007, 95, 163–318. [Google Scholar] [CrossRef]
- Exploiting Genetic Variability of Resistance Genes in Major European Food Legumes to Improve Varieties For Sustainable Agriculture; ERA-PG Funded Project; GENXPro GmbH: Frankfurt, Germany, 2010. Available online: http://www.erapg.org/publicitem.m?key=everyone&pgid=19083&trail=/everyone/16790/18613/18624/19083 (accessed on 12 March 2012).
- Kahl, G.; Winter, P.; Horres, R.; Rotter, B.; Jüngling, R.; Consortium, L. Pathogenesis-related genes and genetic variation in potential resistance genes of major European legumes: The LEGRESIST project. In Ascochyta 2009: Proceedings of the Second International Ascochyta Workshop, Pullman, WA, USA, 28 June–2 July 2009; p. 47.
- Goodman, M.M. Plant breeding requirements for applied molecular biology. Crop Sci. 2004, 44, 1913–1914. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gnanasambandam, A.; Paull, J.; Torres, A.; Kaur, S.; Leonforte, T.; Li, H.; Zong, X.; Yang, T.; Materne, M. Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies. Agronomy 2012, 2, 132-166. https://doi.org/10.3390/agronomy2030132
Gnanasambandam A, Paull J, Torres A, Kaur S, Leonforte T, Li H, Zong X, Yang T, Materne M. Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies. Agronomy. 2012; 2(3):132-166. https://doi.org/10.3390/agronomy2030132
Chicago/Turabian StyleGnanasambandam, Annathurai, Jeff Paull, Ana Torres, Sukhjiwan Kaur, Tony Leonforte, Haobing Li, Xuxiao Zong, Tao Yang, and Michael Materne. 2012. "Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies" Agronomy 2, no. 3: 132-166. https://doi.org/10.3390/agronomy2030132
APA StyleGnanasambandam, A., Paull, J., Torres, A., Kaur, S., Leonforte, T., Li, H., Zong, X., Yang, T., & Materne, M. (2012). Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies. Agronomy, 2(3), 132-166. https://doi.org/10.3390/agronomy2030132




