QTL for Water Use Related Traits in Juvenile Barley
Abstract
:1. Introduction
2. Results
2.1. Phenotyping
2.2. Genome Wide Association Study
3. Discussion
4. Materials and Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shao, H.-B.; Chu, L.-Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.-A. Understanding water deficit stress-induced changes in the basic metabolism of higher plants–biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechol. 2009, 29, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, S.; Sawant, K. Drought stress adaptation: Metabolic adjustment and regulation of gene expression. Plant Breed. 2013, 132, 21–32. [Google Scholar] [CrossRef]
- Barnabas, B.; Jager, K.; Feher, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.A.; Xie, X.-Y.; Wang, L.-C.; Saleem, M.F.; Man, C.; Lei, W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 2011, 6, 2026–2032. [Google Scholar]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Reynolds, M.; Mujeeb-Kazi, A.; Sawkins, M. Prospects for utilising plant-adaptive mechanisms to improve wheat and other crops in drought-and salinity-prone environments. Ann. Appl. Biol. 2005, 146, 239–259. [Google Scholar] [CrossRef]
- El Hafid, R.; Smith, D.H.; Karrou, M.; Samir, K. Physiological responses of spring durum wheat cultivars to early-season drought in a mediterranean environment. Ann. Bot. 1998, 81, 363–370. [Google Scholar] [CrossRef]
- Blum, A. Osmotic adjustment and growth of barley genotypes under drought stress. Crop Sci. 1989, 29, 230–233. [Google Scholar] [CrossRef]
- Fricke, W.; Pritchard, J.; Leigh, R.A.; Tomos, A.D. Cells of the upper and lower epidermis of barley (Hordeum vulgare L.) leaves exhibit distinct patterns of vacuolar solutes. Plant Physiol. 1994, 104, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Tyerman, S.; Niemietz, C.; Bramley, H. Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002, 25, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Kosova, K.; Vitamvas, P.; Urban, M.O.; Kholova, J.; Prášil, I.T. Breeding for enhanced drought resistance in barley and wheat-drought-associated traits, genetic resources and their potential utilization in breeding programmes. Czech J. Genet. Plant Breed. 2014, 50, 247–261. [Google Scholar]
- Honsdorf, N.; March, T.J.; Hecht, A.; Eglinton, J.; Pillen, K. Evaluation of juvenile drought stress tolerance and genotyping by sequencing with wild barley introgression lines. Mol. Breed. 2014, 34, 1475–1495. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, H.; Dai, H.; Zhang, G.; Wu, F. Difference in response to drought stress among tibet wild barley genotypes. Euphytica 2010, 172, 395–403. [Google Scholar] [CrossRef]
- Weatherley, P. Studies in the water relations of the cotton plant. New Phytol. 1950, 49, 81–97. [Google Scholar] [CrossRef]
- Vadez, V.; Kholova, J.; Medina, S.; Kakkera, A.; Anderberg, H. Transpiration efficiency: New insights into an old story. J. Exp. Bot. 2014, 65, 6141–6153. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, G.; Richards, R. Isotopic composition of plant carbon correlates with water-use efficiency of wheat genotypes. Funct. Plant Biol. 1984, 11, 539–552. [Google Scholar] [CrossRef]
- Farquhar, G.D.; Ehleringer, J.E.; Hubick, K.T. Carbon isotope discrimination and photosynthesis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 503–537. [Google Scholar] [CrossRef]
- Boyer, J.S. Advances in drought tolerance in plants. Adv. Agron. 1996, 56, 187–219. [Google Scholar]
- Tuberosa, R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Morison, J.I.; Baker, N.R.; Mullineaux, P.M.; Davies, W.J. Improving water use in crop production. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 639–658. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Effective use of water (EUW) and not water-use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crop Res 2009, 112, 119–123. [Google Scholar] [CrossRef]
- Condon, A.G.; Richards, R.A.; Rebetzke, G.J.; Farquhar, G.D. Improving intrinsic water-use efficiency and crop yield. Crop Sci. 2002, 42, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Condon, A.G.; Richards, R.; Rebetzke, G.; Farquhar, G. Breeding for high water-use efficiency. J. Exp. Bot. 2004, 55, 2447–2460. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Available online: http://faostat3.Fao.Org/faostat-gateway/go/to/home/e (accessed on 13 November 2015).
- Von Bothmer, R.; van Hintum, T.; Knüpffer, H.; Sato, K. Diversity in Barley (Hordeum vulgare); Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance qtl in wild barley introgression lines. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S.; James, R.A.; Munns, R.; Condon, T.A.; Passioura, J.B. Osmotic adjustment leads to anomalously low estimates of relative water content in wheat and barley. Funct. Plant Biol. 2008, 35, 1172–1182. [Google Scholar] [CrossRef]
- Hubick, K.; Farquhar, G. Carbon isotope discrimination and the ratio of carbon gained to water lost in barley cultivars. Plant Cell Environ. 1989, 12, 795–804. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- McAusland, L.; Davey, P.; Kanwal, N.; Baker, N.R.; Lawson, T. A novel system for spatial and temporal imaging of intrinsic plant water use efficiency. J. Exp. Bot. 2013, 64, 4993–5007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackay, T.F.; Stone, E.A.; Ayroles, J.F. The genetics of quantitative traits: Challenges and prospects. Nat. Rev. Genet. 2009, 10, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Ganal, M.W.; Polley, A.; Graner, E.-M.; Plieske, J.; Wieseke, R.; Luerssen, H.; Durstewitz, G. Large SNP arrays for genotyping in crop plants. J. Biosci. 2012, 37, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Comadran, J.; Kilian, B.; Russell, J.; Ramsay, L.; Stein, N.; Ganal, M.; Shaw, P.; Bayer, M.; Thomas, W.; Marshall, D.; et al. Natural variation in a homolog of antirrhinum centroradialis contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Silvar, C.; Martis, M.M.; Nussbaumer, T.; Haag, N.; Rauser, R.; Keilwagen, J.; Korzun, V.; Mayer, K.F.; Ordon, F.; Perovic, D. Assessing the barley genome zipper and genomic resources for breeding purposes. Plant Genome J. 2015, 8. [Google Scholar] [CrossRef]
- Varshney, R.K.; Paulo, M.J.; Grando, S.; van Eeuwijk, F.A.; Keizer, L.C.P.; Guo, P.; Ceccarelli, S.; Kilian, A.; Baum, M.; Graner, A. Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Field Crop Res. 2012, 126, 171–180. [Google Scholar] [CrossRef]
- Teulat, B.; Borries, C.; This, D. New qtls identified for plant water status, water-soluble carbohydrate and osmotic adjustment in a barley population grown in a growth-chamber under two water regimes. Theor. Appl. Genet. 2001, 103, 161–170. [Google Scholar] [CrossRef]
- Diab, A.A.; Teulat-Merah, B.; This, D.; Ozturk, N.Z.; Benscher, D.; Sorrells, M.E. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor. Appl. Genet. 2004, 109, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chang, S.X.; Anyia, A.O. Quantitative trait loci for water-use efficiency in barley (Hordeum vulgare L.) measured by carbon isotope discrimination under rain-fed conditions on the canadian prairies. Theor. Appl. Genet. 2012, 125, 71–90. [Google Scholar] [CrossRef] [PubMed]
- Wehner, G.; Balko, C.; Enders, M.; Humbeck, K.; Ordon, F. Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, P.D.; Martin, R.J.; Francis, G.S.; Wilson, D.R. Drought effects on biomass production and radiation-use efficiency in barley. Field Crop Res. 1995, 43, 77–86. [Google Scholar] [CrossRef]
- Samarah, N.H. Effects of drought stress on growth and yield of barley. Agron. Sustain. Dev. 2005, 25, 145–149. [Google Scholar] [CrossRef]
- Richards, R.; Rebetzke, G.; Condon, A.; Van Herwaarden, A. Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci. 2002, 42, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Selection for sustained production in water-deficit environments. In International Crop Science I; Buxton, D., Ed.; CSSA: Madison, WI, USA, 1993; pp. 343–347. [Google Scholar]
- Ahmed, I.M.; Dai, H.; Zheng, W.; Cao, F.; Zhang, G.; Sun, D.; Wu, F. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between tibetan wild and cultivated barley. Plant Physiol. Biochem. 2013, 63, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Robredo, A.; Pérez-López, U.; de la Maza, H.S.; González-Moro, B.; Lacuesta, M.; Mena-Petite, A.; Muñoz-Rueda, A. Elevated CO2 alleviates the impact of drought on barley improving water status by lowering stomatal conductance and delaying its effects on photosynthesis. Environ. Exp. Bot. 2007, 59, 252–263. [Google Scholar] [CrossRef]
- Blum, A. Drought resistance, water-use efficiency, and yield potential—Are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 56, 1159–1168. [Google Scholar] [CrossRef]
- Sivamani, E.; Bahieldin, A.; Wraith, J.M.; Al-Niemi, T.; Dyer, W.E.; Ho, T.-H.D.; Qu, R. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley hva1 gene. Plant Sci. 2000, 155, 1–9. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, Y.; Breeze, E.; Koo, J.C.; Woo, H.R.; Ryu, J.S.; Park, D.H.; Beynon, J.; Tabrett, A.; Buchanan-Wollaston, V. Overexpression of a chromatin architecture-controlling at-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007, 52, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Munne-Bosch, S.; Alegre, L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 2004, 31, 203–216. [Google Scholar] [CrossRef]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Bourgis, F.; Ziemak, M.J.; Rhodes, D.; Gage, D.A.; Hanson, A.D. Characterization and functional expression of cDNAs encoding methionine-sensitive and-insensitive homocysteine S-methyltransferases from arabidopsis. J. Biol. Chem. 2000, 275, 15962–15968. [Google Scholar] [CrossRef] [PubMed]
- Nohzadeh Malakshah, S.; Habibi Rezaei, M.; Heidari, M.; Hosseini Salekdeh, G. Proteomics reveals new salt responsive proteins associated with rice plasma membrane. Biosci. Biotechnol. Biochem. 2007, 71, 2144–2154. [Google Scholar] [CrossRef] [PubMed]
- Handley, L.L.; Nevo, E.; Raven, J.A.; MartInez-Carrasco, R.; Scrimgeour, C.M.; Pakniyat, H.; Forster, B.P. Chromosome 4 controls potential water use efficiency (δ13c) in barley. J. Exp. Bot. 1994, 45, 1661–1663. [Google Scholar] [CrossRef]
- Molnár, I.; Linc, G.; Dulai, S.; Nagy, E.; Molnár-Láng, M. Ability of chromosome 4h to compensate for 4D in response to drought stress in a newly developed and identified wheat–barley 4H (4D) disomic substitution line. Plant Breed. 2007, 126, 369–374. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Satou, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in arabidopsis. Plant Cell 2005, 17, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, L.; Merchan, F.; Laporte, P.; Thompson, R.; Clarke, J.; Sousa, C.; Crespi, M. A novel plant leucine-rich repeat receptor kinase regulates the response of medicago truncatula roots to salt stress. Plant Cell 2009, 21, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Deepak, S.; Shailasree, S.; Kini, R.K.; Muck, A.; Mithöfer, A.; Shetty, S.H. Hydroxyproline-rich glycoproteins and plant defence. J. Phytopathol. 2010, 158, 585–593. [Google Scholar] [CrossRef]
- Eastburn, D.; McElrone, A.; Bilgin, D. Influence of atmospheric and climatic change on plant–pathogen interactions. Plant Pathol. 2011, 60, 54–69. [Google Scholar] [CrossRef]
- Wehner, G.; Balko, C.; Humbeck, K.; Zyprian, E.; Ordon, F. Expression profiling of genes involved in drought stress and leaf senescence in juvenile barley. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Rode, J.; Ahlemeyer, J.; Friedt, W.; Ordon, F. Identification of marker-trait associations in the german winter barley breeding gene pool (Hordeum vulgare L.). Mol. Breed. 2012, 30, 831–843. [Google Scholar] [CrossRef]
- Igartua, E.; Gracia, M.P.; Lasa, J.M.; Medina, B.; Molina-Cano, J.L.; Montoya, J.L.; Romagosa, I. The spanish barley core collection. Genet. Resour. Crop Evolut. 1998, 45, 475–481. [Google Scholar] [CrossRef]
- Wehner, G.; Balko, C.; Ordon, F. Experimental design to determine drought stress response and early leaf senescence in barley (Hordeum vulgare L.). Bio-Protocol 2016, 6. [Google Scholar] [CrossRef]
- Stauss, R. Compendium of Growth Stage Identification Keys for Mono-and Dicotyledonous Plants: Extended bbch Scale; Ciba-Geigy AG: Basel, Switzerland, 1994. [Google Scholar]
- Grubbs, F.E. Procedures for detecting outlying observations in samples. Technometrics 1969, 11, 1–21. [Google Scholar] [CrossRef]
- RStudio Team. Rstudio: Integrated development for r. Available online: http://www.rstudio.com/ (accessed on 13 June 2016).
- Reif, J.C.; Melchinger, A.E.; Frisch, M. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci. 2005, 45, 1–7. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]
- Galaxy. Available online: https://galaxyproject.org/ (accessed on 11 July 2016).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Tassel 5.0. Available online: http://www.maizegenetics.net/tassel (accessed on 25 July 2016).
- Mayer, K.F.; Waugh, R.; Brown, J.W.; Schulman, A.; Langridge, P.; Platzer, M.; Fincher, G.B.; Muehlbauer, G.J.; Sato, K.; Close, T.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. Barlex—The barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef] [PubMed]
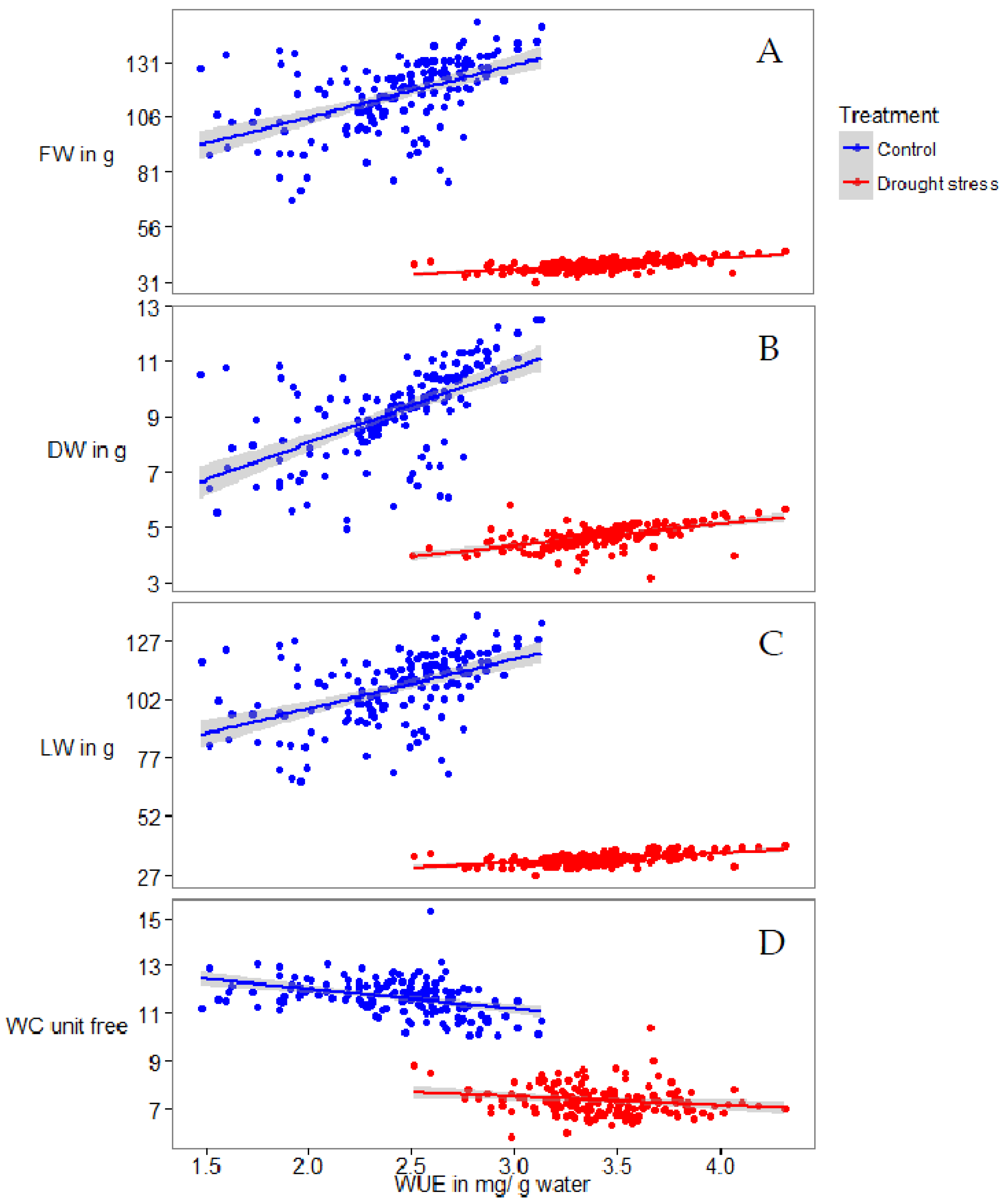
| Trait a | Description | Calculation | Treat. b | Unit | Min c | Max c | Mean c | SD c | CV c | LSD c |
|---|---|---|---|---|---|---|---|---|---|---|
| FW | Fresh weight of shoot | Control | g | 67.96 | 188.00 | 120.92 | 19.18 | 0.16 | 27.86 | |
| biomass per pot | Stress | g | 26.33 | 45.33 | 36.62 | 3.35 | 0.09 | 7.23 | ||
| LW | Total leaf water | FW-DW | Control | g | 64.20 | 177.83 | 111.81 | 17.66 | 0.16 | 26.11 |
| Stress | g | 23.27 | 39.68 | 32.32 | 3.03 | 0.09 | 6.73 | |||
| WC | Leaf water content | LW/DW | Control | unit free | 10.06 | 17.15 | 12.24 | 1.02 | 0.08 | 2.03 |
| Stress | unit free | 5.69 | 12.17 | 7.60 | 1.06 | 0.14 | 2.20 | |||
| WUE | Water use efficiency | DW/total water added | Control | mg/g water | 1.48 | 3.13 | 2.42 | 0.34 | 0.14 | 0.58 |
| Stress | mg/g water | 2.51 | 4.31 | 3.42 | 0.31 | 0.09 | 1.11 | |||
| Trait a | Effect b | F Value | p Value |
|---|---|---|---|
| FW | Genotype | 5.12 | <0.0001 |
| Treatment | 18,938.63 | <0.0001 | |
| Year | 39.38 | <0.0001 | |
| G × T | 4.01 | <0.0001 | |
| G × Y | 1.51 | 0.0002 | |
| LW | Genotype | 5.11 | <0.0001 |
| Treatment | 20,443.78 | <0.0001 | |
| Year | 41.61 | <0.0001 | |
| G × T | 4.21 | <0.0001 | |
| G × Y | 1.50 | 0.0002 | |
| WC | Genotype | 4.71 | <0.0001 |
| Treatment | 8631.12 | <0.0001 | |
| Year | 245.29 | <0.0001 | |
| G × T | 1.35 | 0.0048 | |
| G × Y | 2.39 | <0.0001 | |
| WUE | Genotype | 3.89 | <0.0001 |
| Treatment | 1928.18 | <0.0001 | |
| G × T | 1.35 | 0.0076 |
| Trait a | Treatment | DW | LW | WC | WUE |
|---|---|---|---|---|---|
| FW | Control | 0.92 *** | 1 *** | −0.26 ** | 0.55 *** |
| Drought stress | 0.59 *** | 0.98 *** | 0.18 * | 0.59 *** | |
| DW | Control | 0.91 *** | −0.54 *** | 0.64 *** | |
| Drought stress | 0.48 *** | −0.60 *** | 0.65 *** | ||
| LW | Control | −0.22 ** | 0.54 *** | ||
| Drought stress | 0.31 ** | 0.54 ** | |||
| WC | Control | −0.44 *** | |||
| Drought stress | −0.22 ** |
| Trait a | Treat. b | SNP Marker c | Chr. c | Pos. in cM c | F Value | p Value | -log p (LOD)d | R2 in % | Morex Contig e | Conf. e | Functional Annotation e | Gene Name e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | S | i_SCRI_RS_210101 | 3H | 21.38 | 7.10 | 0.0010 | 3.01 | 3.46 | morex_contig_46157 | HC | Eukaryotic translation initiation factor 3 subunit, putative | MLOC_62056.2 |
| WC | C | i_SCRI_RS_150063 | 3H | 113.27 | 8.03 | 0.0004 | 3.40 | 3.73 | morex_contig_6173 | HC | Leucine-rich repeat receptor-like protein kinase | MLOC_72476.1 |
| WC | C | i_SCRI_RS_203905 | 3H | 113.27 | 7.39 | 0.0007 | 3.13 | 3.43 | morex_contig_2551243 | HC | Leucine-rich repeat receptor-like protein kinase | MLOC_38740.7 |
| WC | C | i_11_20527 | 3H | 124.89 | 7.39 | 0.0007 | 3.13 | 3.43 | morex_contig_44198 | HC | Hydroxyproline-rich glycoprotein-like | MLOC_60122.1 |
| WC | C | i_12_30921 | 3H | 147.34 | 12.04 | 0.0006 | 3.22 | 2.79 | morex_contig_36914 | HC | RNA-dependent RNA polymerase | MLOC_51409.1 |
| WUE | S | i_11_10319 | 4H | 14.74 | 7.98 | 0.0005 | 3.29 | 10.50 | morex_contig_2553017 | HC | Pathogenesis-related thaumatin-like protein | MLOC_39318.1 |
| WUE | C | i_12_31385 | 4H | 61.21 | 6.35 | 0.0004 | 3.35 | 12.62 | morex_contig_2548269 | HC | Homocysteine S-methyltransferase 1, putative, expressed | MLOC_37381.1 |
| WUE | C | i_SCRI_RS_151213 | 4H | 61.56 | 5.72 | 0.0010 | 3.00 | 11.36 | morex_contig_276516 | LC | - | MLOC_45156.1 |
| WC | C | i_SCRI_RS_203117 | 5H | 36.18 | 7.23 | 0.0009 | 3.06 | 3.36 | morex_contig_1581968 | LC | - | MLOC_18779.2 |
| LW | C | i_SCRI_RS_220136 | 5H | 80.33 | 13.60 | 0.0003 | 3.57 | 3.84 | morex_contig_1565092 | HC | unknown protein; involved in: N-terminal protein myristoylation | MLOC_13193.1 |
| LW | S | i_11_21247 | 5H | 124.52 | 12.62 | 0.0004 | 3.35 | 2.82 | morex_contig_135323 | HC | Protein kinase, putative | MLOC_4611.1 |
| HC | Aluminum-induced protein-like | MLOC_51340.2 | ||||||||||
| LW | S | i_12_21471 | 5H | 124.52 | 12.62 | 0.0004 | 3.35 | 2.82 | morex_contig_36899 | HC | Transcription factor, putative | AK373628 |
| WUE | C | i_11_21528 | 7H | 47.14 | 9.74 | 0.0001 | 3.97 | 12.89 | morex_contig_1582615 | HC | WD repeat-containing protein, putative | AK366046 |
| LW | C | i_SCRI_RS_202061 | 7H | 48.49 | 8.08 | 0.0004 | 3.41 | 4.56 | morex_contig_1567807 | HC | unknown protein; | AK372377 |
| HC | AT hook motif DNA-binding family protein | MLOC_14326.3 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wehner, G.; Balko, C.; Ordon, F. QTL for Water Use Related Traits in Juvenile Barley. Agronomy 2016, 6, 62. https://doi.org/10.3390/agronomy6040062
Wehner G, Balko C, Ordon F. QTL for Water Use Related Traits in Juvenile Barley. Agronomy. 2016; 6(4):62. https://doi.org/10.3390/agronomy6040062
Chicago/Turabian StyleWehner, Gwendolin, Christiane Balko, and Frank Ordon. 2016. "QTL for Water Use Related Traits in Juvenile Barley" Agronomy 6, no. 4: 62. https://doi.org/10.3390/agronomy6040062
APA StyleWehner, G., Balko, C., & Ordon, F. (2016). QTL for Water Use Related Traits in Juvenile Barley. Agronomy, 6(4), 62. https://doi.org/10.3390/agronomy6040062





