Basic statistics and a wide range (18.0–50.0%) of percent significant pairwise differences (
p < 0.05) between 13 maize genotypes for kernel nutrient densities (mg kg
−1) classified into four HGEs and based on 1348 maize accessions are presented in
Table 1. Mean macronutrient density (mg kg
−1) was dominated, in decreasing order, by [K, P, Mg and S] densities, whereas [Fe and Zn] densities dominated for micronutrients. Four macronutrients [K, P, Mg and S], in decreasing order, accounted for 91.0% of total [nutrient] densities in this germplasm. Coefficient of variation (C.V.) was larger for micronutrients (16.0–36.0%) as compared to macronutrients (4.0–16.1%). The Shapiro-Wilk Statistical test of normality (i.e., SW-W was significant for all nutrients, except Mg and P. Two nutrients (N and S) were negatively skewed, while the remaining nutrients were positively skewed. Summary statistics based on ANOVA and mean separation (DNMRT;
p < 0.05) indicated that more significant pairwise differences for macronutrients (32.3%) existed between genotypes as compared to micronutrients (26.5%) or the FeZn-Index (23.0%). The largest (50%) and smallest (23%) values were recorded between genotypes for [K and C] densities, respectively, whereas the respective values for micronutrients were 35% for Mn and 19% for Zn densities. The M-Index (0.78 ± 0.05; C.V. = 6.0%) differed (
p < 0.05) from the m-Index (0.61 ± 0.07; C.V. = 11.0%), and from the FeZn-Index (0.63.0 ± 0.07; C.V. = 11.0%).
3.1. Variance Components
The effects of fixed and random factors and covariates on macro-and micronutrients are shown in
Table 2. The univariate significant (
p < 0.05) R
2 estimates due to fixed factors (i.e., HG, E, IB, and G) on macronutrients ranged from 1.8% of E on Mg to 56.9% of G on K density; the respective values for micronutrients were 1.5% for HG on Mn and 54.4% for G on Cu density. The only non-significant effects were those for HG for all macronutrients, and E for both Ca and S densities. The effects of IB and G on macro- and micronutrients were, in general, larger than those of HG and E. Significant (z < 0.05) total variation explained by random factors (i.e., nested E, G, IB within HG; and G × IB nested within HG) ranged from 45.3% S to 76.7% K density for macronutrients, and from 50.8% Mn to 70.1% Cu density for micronutrients. The largest percent variation explained by E(HG) in macronutrients was 12.2% for Mg density and in micronutrients it was 16.2% for Cu density. The respective values for G(HG) were 46.3% K and 44.2% Cu densities, for IB(HG) the respective values were 31.2% N and 15.2% Fe, and for G × IB(HG) these values were 47.7 P, and 42.6% Mn densities.
Significant covariate effects (R
2) on macro- and micronutrients were presented (
Table 2) if linear (β1), quadratic (β2), or both regression coefficients were significant (
p < 0.05). Biochemical constituents of the maize kernel had positive (32%) or negative (32%) linear effects on macronutrients, while 23% and 11% of their quadratic effects were positive and negative, respectively. The largest negative effects on macronutrients were those of the C:N ratio, followed, in decreasing order, by starch and K-moisture. Starch and C:N ratio, in particular, had negative linear and quadratic effects on [Ca and N], respectively. Protein and amino acid contents generally had positive effects on most macronutrients; whereas oil content had the smallest negative effects on a few [Ca, N and S] macronutrients.
Kernel biochemicals had almost equal positive (28%) and negative linear effects (29%) on micronutrients. Protein, amino acid (lysine, methionine, cysteine), and oil contents had positive effects on all micronutrients, while the remaining biochemicals had negative linear effects. Kernel physical attributes mainly had negative effects on all macronutrients, except N, while both negative linear and positive quadratic effects were observed for micronutrients. Kernel density was the only attribute with significant negative linear and quadratic effects on all micronutrients. None of the kernel physical attributes affected S estimates, while estimates of Fe, Mn and Zn were not affected by K-volume.
All fixed factors had significant effects on the M-Index and m-Index, as indicated by significant R
2 estimates. Both G and IB, in decreasing order, had larger effects on both indices than HG and E. All fixed factors, except E, had significant effects on the FeZn-Index; while all random factors accounted for some variation in each of the three indices, with two exceptions (
Table 2). The largest portion (41.3%) of total variation (76.0%) in the FeZn-Index was attributed to G × IB(HG); it was larger than variation explained in the M-Index (52.4%), or the m-Index (55.9%). However, effects of individual random factors differed among these indices. The G × IB(HG) accounted for the most variation in the M-Index and FeZn-Index, while G(HG) accounted for most of the variation in the m-Index.
Covariate effects on all indices mirrored, to a large extent, their effects on the respective nutrients used in their estimation. The C:N ratio had a significant negative effect on all indices; protein and amino acids had positive linear and quadratic effects on all three indices with large R2 estimates; and starch and K-moisture had parallel and significant negative linear and positive quadratic effects on all but the FeZn-Index. K-density was the only physical trait with negative linear and quadratic effects on all indices, while oil had a negligible positive effect on the m-Index.
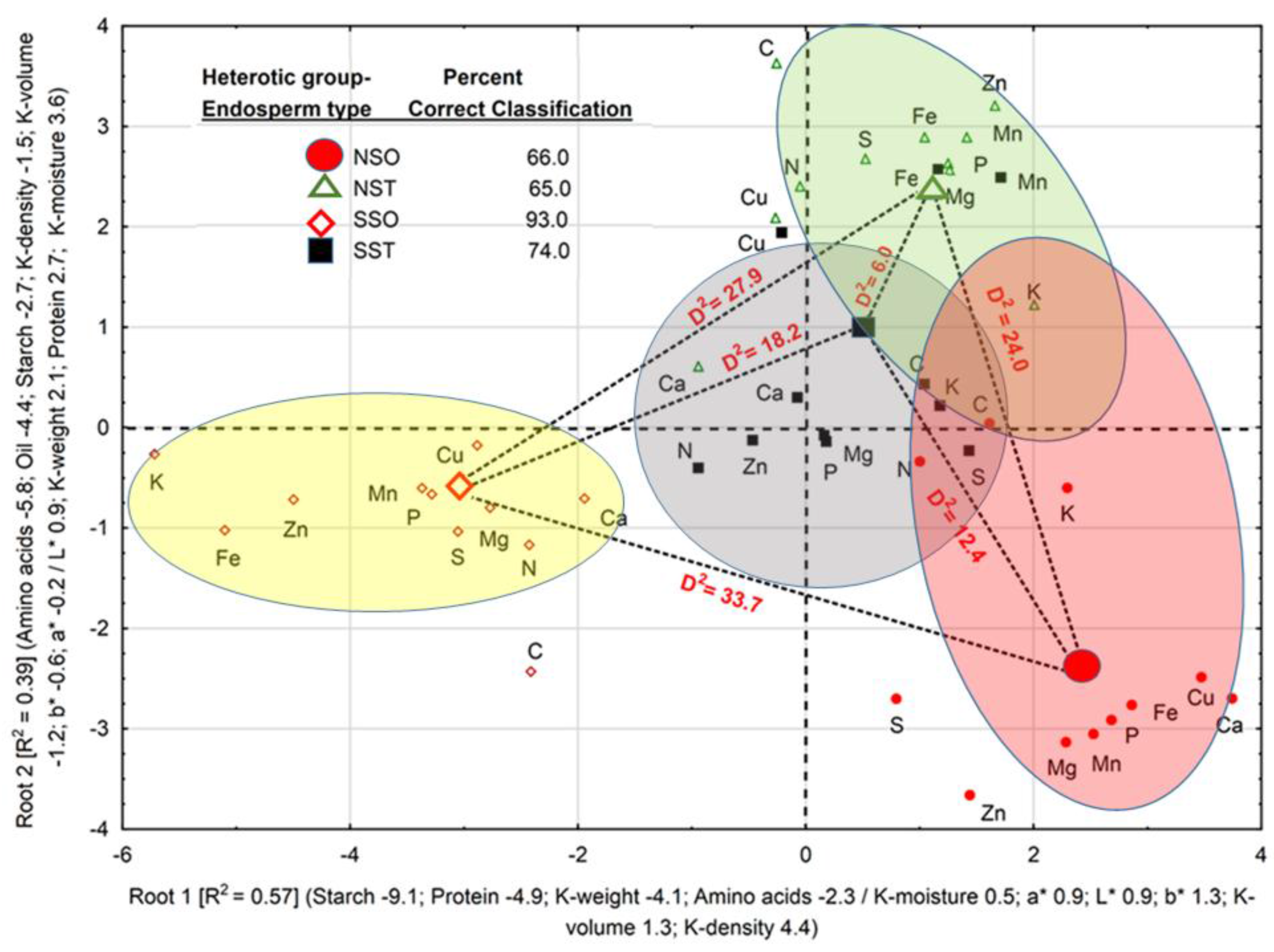
Physical, biochemical, and color space coordinates discriminated between HGEs with 74.5% correct classification, and in two discriminant roots, accounted for 96.0% of total variation (
Figure 1). Percent correct classifications of HGEs were 66.0 (NSO), 65.0 (NST), 93.0 (SSO), and 74.0% (SST). Positive loadings of K-density, K-volume,
b*,
L*,
a*, and K-moisture, in decreasing order, and negative loadings of starch, protein, K-weight and amino acids, in decreasing order, accounted for 57.0% of total variation and separated the SSO from the remaining HGEs along Root 1, while Root 2 accounted for 39.0% of total variation and separated the SSO and NSO from the SST and NST. All D
2 values were significant (
p < 0.05); the largest D
2 (33.0) was found between the NSO and SSO, followed, in decreasing order, by D
2 between the NST and the SSO (27.9), between the NST and NSO (24.0), between the SST and SSO (18.2), between the SST and NSO (12.4), and between the NST and SST (6.0).
The association in each root between centroids and loadings of physical attributes, biochemical constituents, and nutrients, suggested that accessions within the SSO have larger values for those attributes and constituents with negative loadings on Root 1 and Root 2. Accessions of the NSO, NST, and SST, in decreasing order, have larger values of kernel attributes and constituents with positive loadings on Root 1. The same reasoning can be applied to loadings on Root 2. Nutrients in accessions within each HGE clustered around its centroid in accordance with its percent correct classification. The association of a few nutrients (e.g., [C and K] in NST, NSO and SST) with their HGEs centroids was weaker than most nutrients; thus contributing to smaller percent correct classification of HGEs.
3.2. Single and Multiple Nutrient Associations
Variance (R
2) explained by kernel physical, biochemical, and color space coordinates, in the M-Index and the m-Index for each genotype with OE or TE and classified into the SS or NS heterotic groups, ranged from 0.001 to 0.63 (
Figure 2). Four clusters (
p < 0.05) were delineated at the 40% linkage distance (
Y-axis). Cluster (1) was composed of amino acid, starch, and protein values, cluster (2) joined oil and
b* values, cluster (3) was composed of K-moisture, K-density, and K-volume values, and cluster (4) joined K-weight,
L*, and
a* values. The 50 combinations of genotypes-indices-endosperm textures were separated into several color-coded clusters based on similarities of R
2 estimates (
X-axis). The 40% linkage distance delineated six (
p < 0.05) clusters; the smallest (4) was composed of the genotype LH119-AR16035, while the largest (6) had 23 combinations of M-Index and m-Index under HGE. The clustering procedure resulted in uniform or mixed grouping of genotypes within HGEs. Both of the genotypes (GQ-N16-N12 and Nokomis Gold) formed uniform clusters across HGs, but not E types. Others were separated among two (e.g., NG-Mo508-Mo506), three (e.g., 1895-DK212-N11a), or four clusters (e.g., DKXL370-S11). Clustering distance (from the origin) was the largest for GQ-N16-N12 and the smallest for two genotypes (1895-DK212-N11a and AR16021-B73), while LH119-AR16035, for example, formed a single cluster. The hierarchical classification of genotypes within HGEs and results of statistical tests indicated that the percentages of significant R
2 values (>5%) for both indices were equal (48.0%). However, slight differences were found between R
2 (
p < 0.05) values of OE (51.0%) and TE (44.0%) textures, and between the NS (49.0%) and SS (46.0%) HGs. The largest differences, however, were attributed to protein, amino acids, and starch with the largest (>0.96%) R
2, followed, in decreasing order, by K-moisture (54.0%) and K-weight (42.0%), oil% (38.0%),
b* (30.0%), K-density (26.0%), K-volume (0.20%), and then
L* and
a* with 10.0% each.
Box-plots and mean separation between M-indices (
Figure 3A), m-Indices (
Figure 3B), and FeZn-Indices (
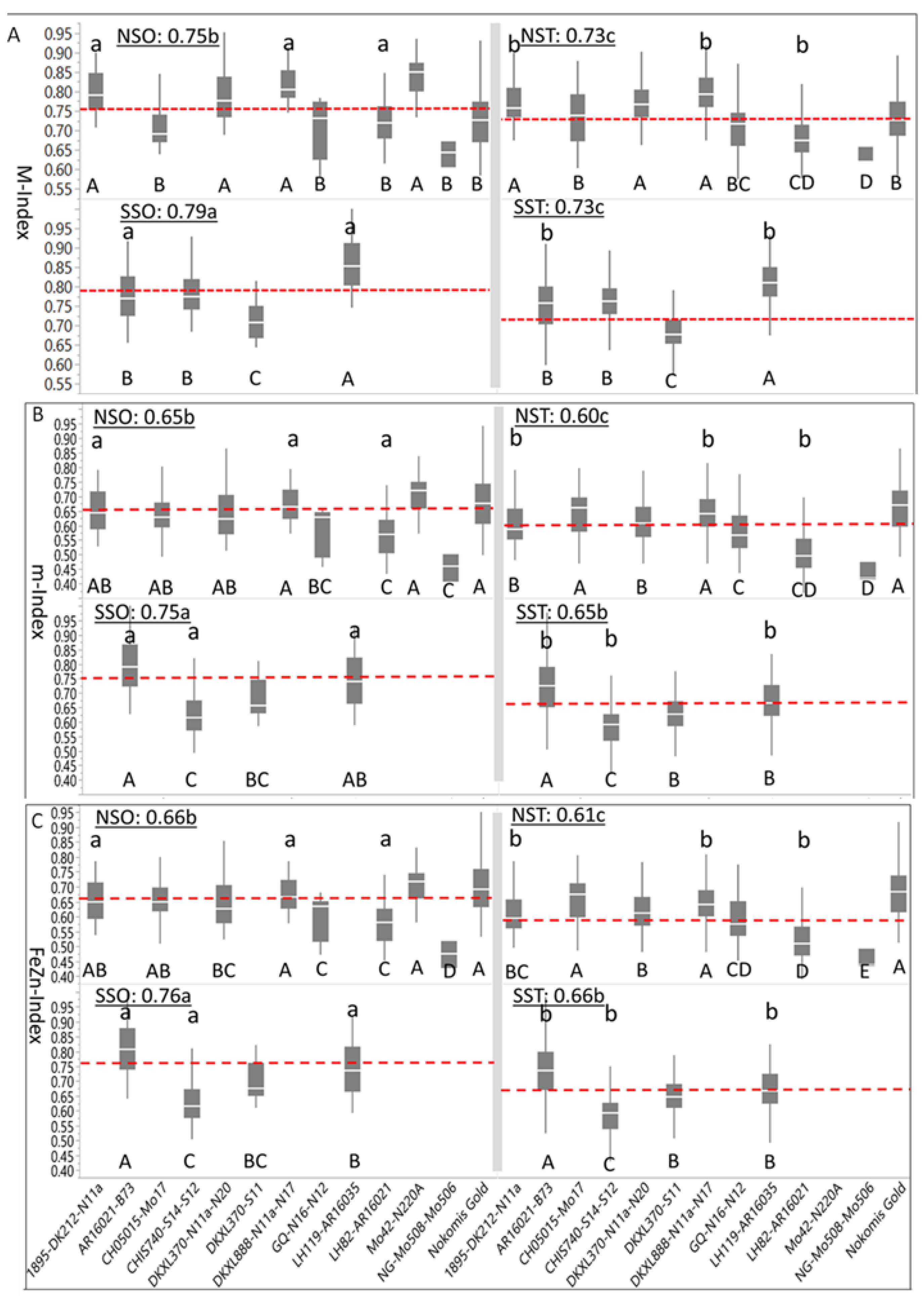
Figure 3C) were based on normalized [nutrient] density in maize kernels. Summary statistics indicated that the overall Index (0.75 ± 0.08) was similar in magnitude and level of variation to the M-Index (0.76 ± 0.08); however, the m-Index (0.65 ± 0.11) was smaller in magnitude and larger in variation, but closely similar to FeZn-Index (0.66 ± 0.11). When averaged over genotypes, the positive skewness of the M-Index (0.15 ± 0.07), m-Index (0.24 ± 0.07), and FeZn-Index (0.26 ± 0.07) was confirmed; however, when HGEs were considered, the m-Index and FeZn-Index of the SSO were negatively skewed, while all other indices of the HGEs were positively skewed. On the other hand, all indices of four genotypes of the NSO (DKXL888-N11a-N17, GQ-N16-N12, Mo42-N220A, and NG-Mo508-Mo506), as well as all indices of two genotypes of the NST (CH05015-Mo17 and DKXL370-S11), and of one genotype (LH119-AR16035) of the SST were negatively skewed.
The weighted average of the M-Index for all genotypes and HGE groups was 0.74 ± 0.08. The SSO genotypes had the largest mean M-Index (0.79), differing (
p < 0.05) from the following M-Index of the NSO (0.75), which also differed (
p < 0.05) from M-Indices of both the NST (0.73) and the SST (0.73) (
Figure 3A). Mean M-Indices of four and six genotypes for the NSO and NST were above the average of their respective HGE means, respectively, whereas one and three genotypes had above average M-Indices for the SSO and SST, respectively. Mean M-Indices for individual genotypes of the NSO were separated into two (
p < 0.05) groups as indicated by capital letters below their boxplots (DNMRT;
p < 0.05), whereas their NST counterparts were separated into five (
p < 0.05) groups. The number of (
p < 0.05) groups of genotypes was three each for SSO and SST. Genotypes in the SSO, with the largest M-Index (0.79), differed (
p < 0.05) from their translucent counterparts (SST) which had the smallest M-Index (0.73). Three NSO genotypes (1895-DK212-N11a, DKXL888-N11a-N17 and LH82-AR16021) had larger (
p < 0.05) M-Indices than their NST counterparts, whereas the same was observed for AR16021-B73 and LH119-AR16021 in the SSO as compared to their SST counterpart.
The weighted average of the m-Index for all genotypes and HGEs was 0.65 ± 0.11; the mean m-Index of SSO (0.75) was larger (
p < 0.05) than for NSO and SST (0.65), while the m-index of NST (0.60) was (
p < 0.05) smaller than the rest (
Figure 3B). Genotypes of the NSO and NST were separated into four and five (
p < 0.05) groups, respectively, whereas all four genotypes within each of the SSO and SST groups differed (
p < 0.05) from each other. Differences between three genotypes of NSO (1895-DK212-N11a, DKXL888-N11a-N17 and LH82-AR16021) from their NST counterparts were significant, while all genotypes but one (DKXL370-S11) in the SSO differed (
p < 0.05) from their SST counterparts.
Numerically, descriptive statistics of the FeZn-Index (
Figure 3C) resembled those of the m-Index. Summary statistics indicated that the weighted average of FeZn-Index for all genotypes and HGE groups was 0.66 ± 0.11. The mean FeZn-Index of SSO was the largest (0.76) and differed (
p < 0.05) from SST (0.66) and NST (0.66), both of which differed significantly from NST (0.61). Genotypes of NSO and NST were separated into five groups each (
p < 0.05), whereas all four genotypes within SSO and SST differed from each other (
p < 0.05). Three genotypes for NSO (1895-DK212-N11a, DKXL888-N11a-N17, and LH82-AR16021) and for SSO (AR16021-B73, CHIS740-S14-S12, and LH119-AR16035) differed (
p < 0.05) from their NST and STT counterparts, respectively.
3.3. Modeling Effect of C:N Allocation
Variance (adjusted R
2) estimates due to β1 and β2 values of C:N on macro- and micronutrients and their indices for HGEs ranged from 0.0 for Cu in SSO to >0.90 for S densities (
Table 3). All β1 values were negative (
p < 0.05) except for Cu in SSO and [Cu and K] densities in SST, whereas all β2 values, regardless of OE or TE texture, were negative for SS and positive for NS. Both sets were significant (
p < 0.05), except for Fe and Zn in the SST, and Ca densities in NS, regardless of OE or TE texture. Most β1 values differed (
t < 0.05) from zero, except [Cu and K]. Meanwhile, β2 values for [Ca, Cu, Mg, Mn, P, K, S, and Zn] densities, as well as those for the M-Index and m-Index, did not (
t > 0.05). In addition, R
2 estimates for [Cu and Fe] densities, and m-Index were not different (
t > 0.05) from zero, while all R
2 estimates differed (
t < 0.05) from 1.0.
Summary statistics suggested that the overall “Mean” and “Response” estimates were positively correlated (r > 0.90; p < 0.001) and, on average, both were negatively correlated with β1 (r = −0.66; p < 001). Also, S.D. and RMSE were correlated (r = 0.88; p < 0.001); S.D. was only correlated with β1 (r = −0.57; p < 0.001), while RMSE was equally correlated with β1 and β2 (−0.34; p < 0.02). Both coefficients were negatively correlated (r = −0.51; p < 0.001), hence the negative (p < 0.05) correlation between β1 (r = −0.43; p < 0.05), but not β2 (r = 0.24; p > 0.05) with R2.
In addition, differences (p < 0.002) in R2 were found between SS (0.34) and NS (0.59), but not between OE (0.51) and TE (0.43) textures (p < 0.38). However, when HGEs were considered, the R2 for NST (0.62) and NSO (0.57) differed (p < 0.01) from SST (0.24), but not from SSO (0.44), which in turn did not differ from NSO or NST. The R2 estimates for genotypes ranged from 0.09 for Cu, to 0.91 for S. Three nutrients had R2 values between 0.60 and 0.66 [Mg, P, and Mn], while Ca (0.46) and Zn (0.42) R2 values were intermediate. The R2 estimates for M-Index (0.48) and m-Index (0.38) did not differ significantly from each other. However, at the genotypic levels, estimates differed significantly (p < 0.01) between SS and NS, respectively for RMSE in Cu, (0.51 and 0.37) and β2 (0.0012 and 0.0035); in Fe for RMSE (4.00 and 2.68), β1 (−0.30 and −0.65), and R2 (0.09 and 0.47); in S for RMSE (54.5 and 36.1); and in Zn for RMSE (2.76 and 1.75), β1 (−0.28 and −0.64), β2 (−0.005 and 0.17), and R2 (0.19 and 0.65). Also, m-Index estimates in SS and NS, respectively, differed (p < 0.01) for RMSE (0.095 and 0.060), β1 (−0.008 and −0.020), and R2 (0.15 and 0.60). On the other hand, differences (p < 0.05) between OE and TE textures were limited to S.D. for Ca (54.4 and 43.7), R2 for Cu (0.01 and 0.17), SD for Fe (3.5 and 3.9), and mean K (3680 and 3520). The estimated M-Index and m-Index averages for NS (0.745 ± 0.02; and 0.628 ± 0.04, respectively) were (p < 0.05) smaller than the respective SS (0.763 ± 0.02; and 0.705 ± 0.04) estimates; however, the respective R2 values were not different.
Also, differences were found between mean M-Index and mean m-Index for OE (0.772 ± 0.012 and 0.703 ± 0.04) and the respective TE texture, with the respective means of 0.736 ± 0.012 and 0.631 ± 0.04, while the respective R2 values were not significantly different. However, when HG and OE/TE data were combined, HGE mean M-Indices were ranked as follows: SSO (0.789) > NSO (0.755) > SST (0.737) = NST (0.735), while the ranking of m-Indices was slightly different: SSO (0.730) > SST (0.656) = NSO (0.652) > NST (0.605).
Large differences were found for β1 and β2 of C:N on [nutrients] and their indices in response to OP or IB (
Table 4). All β1 estimates were negative (
p < 0.05), while β2 estimates for S1 and S2 were positive (
p < 0.05) and those for OP, S3 and S4 were positive but not significant (
p > 0.05). Summary statistics indicated that most β1 values (for [Ca, Fe, Mg, Mn, P, K, S and Zn]) differed from zero (
t < 0.001); however, only the β2 value for Zn did not. Averaged over all IB groups, “Mean” and “Response” did not differ significantly from each other; however, those for S.D. (81.8 ± 122.6) and RMSE (55.6 ± 95.8) differed at
p < 0.07. Similarly, on average, RMSE was negatively (−0.66;
p < 0.05) correlated with β1; however, the large correlation for OP and S1 (−0.80;
p < 0.05) dropped to −0.76 for S2, then to −0.67 for S3 (
p < 0.05), and finally to −0.40 (
p > 0.05) for S4.
Average R2 (0.51 ± 0.26) differed (t < 0.05) from zero and from 1.0. All R2 estimates for individual nutrients differed (t < 0.05) from zero, including the smallest (0.12 ± 0.06, for Cu); also, all R2 estimates, except the largest (0.89 ± 0.04, for S), differed (t < 0.05) from 1.0. Averaged over IBs, similar R2 estimates were found for the M-Index (0.50 ± 0.18) and m-Index (0.46 ± 0.21). Comparisons between mean R2 estimates of IBs indicated that three IB groups OP (0.64) = S1 (0.64) = S2 (0.64) differed (p < 0.05) from the remaining two S3 (0.39) = S4 (0.31). The R2 estimates were negatively (p < 0.05) correlated with β1 in S4 (r = −0.64), and in Fe, K, Mg, Mn and P (r ≥ −0.88), and positively (p < 0.05) with β2 in only Cu (r = 0.91) and S (r = 0.88), as well as with the m-Index (r = 0.89).
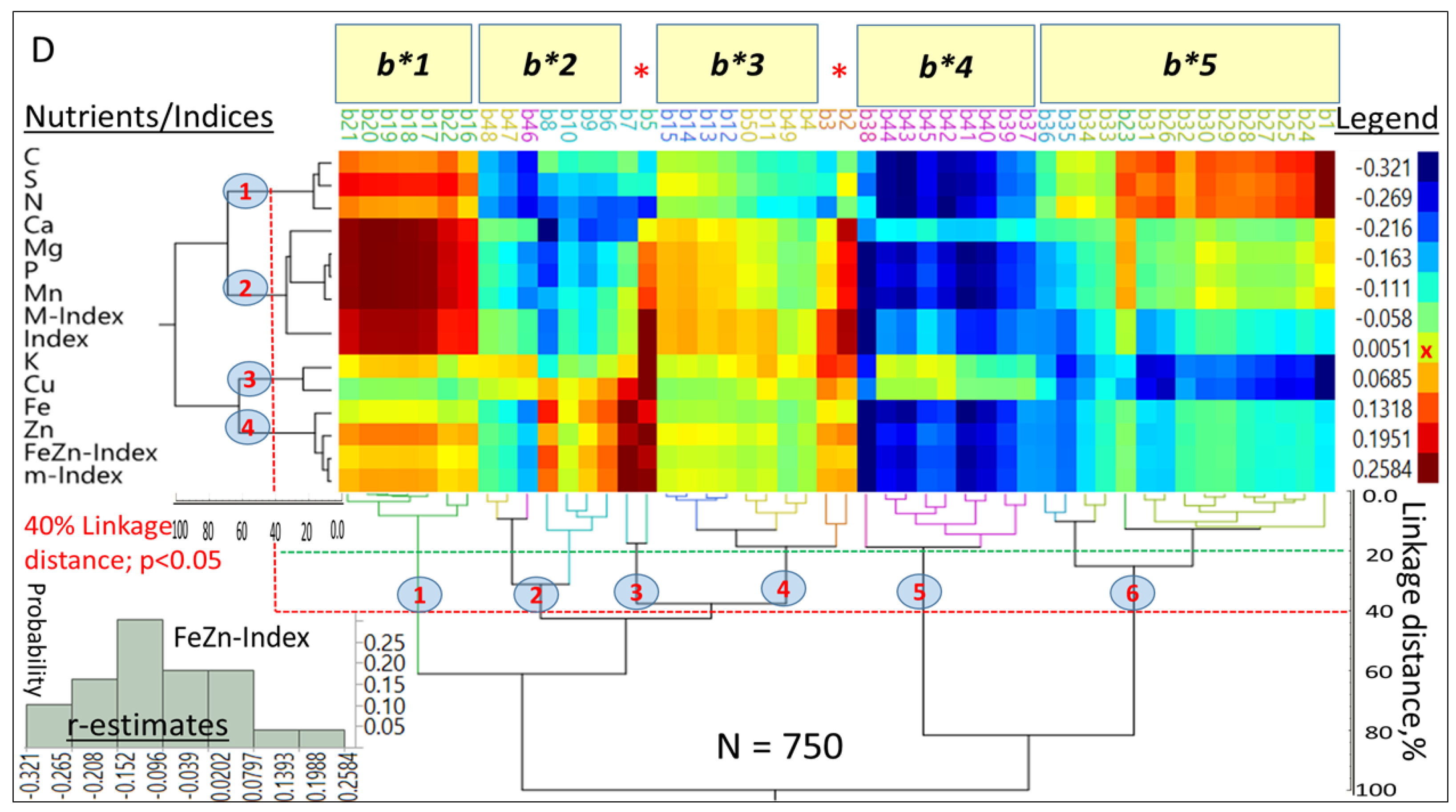
3.4. Modeling Nutrient Indices
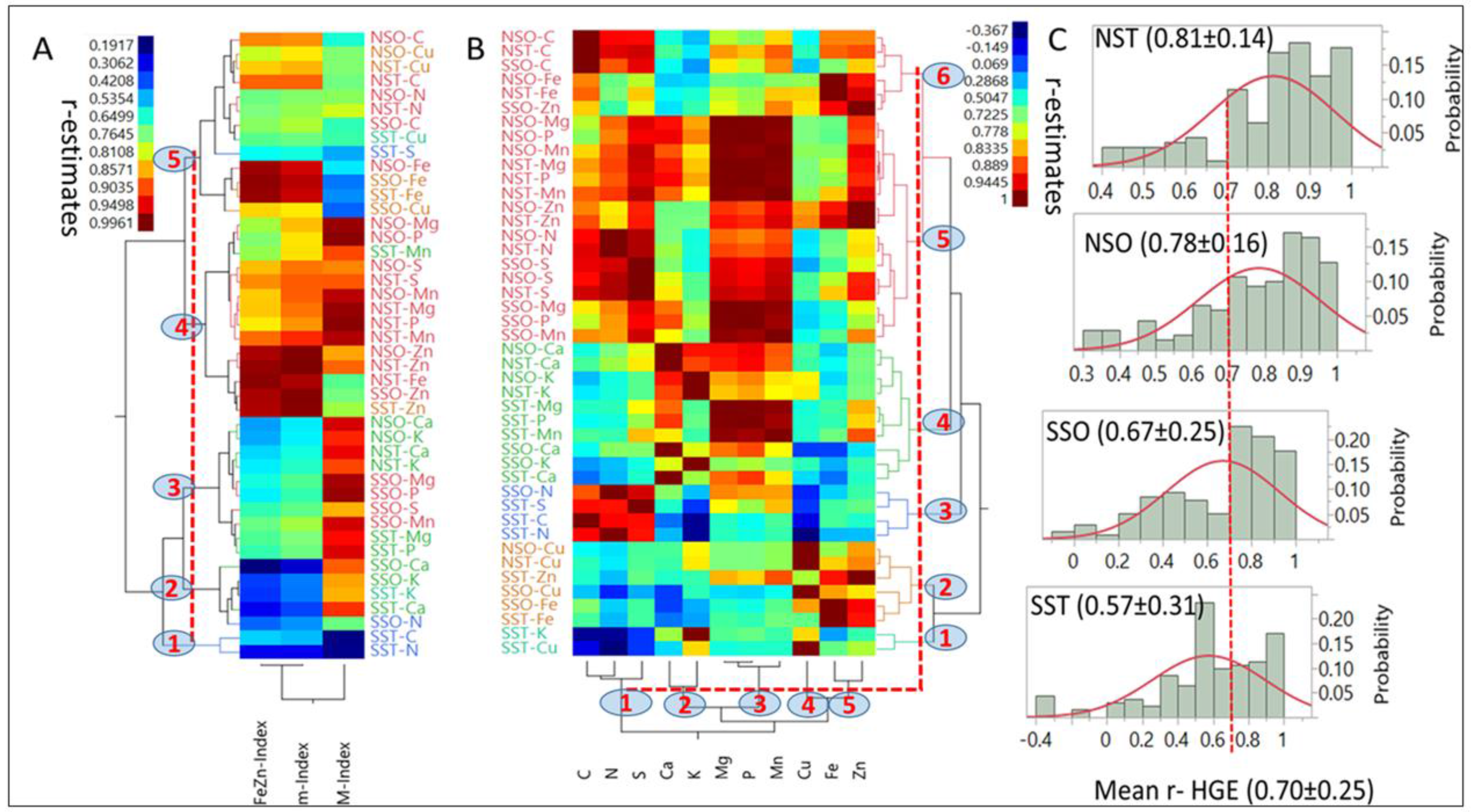
Hierarchical, two-way clustering based on correlation coefficients (r = 0.19 to 0.99;
p < 0.05) between each index and nutrients in each HGEs (
Figure 4A) generated five clusters (
p < 0.05) at the 40% linkage distance. Although mean r-estimates between indices and nutrients were not statistically different (
p > 0.05), there were clear differences in their level of association with individual (e.g., M-Index with C in NSO) or multiple nutrients (e.g., m-Index with [Mg, Mn and P] as compared with their association with the FeZn-Index).
Only Mn had a larger affinity to the M-Index than to the m-Index. Single nutrients exhibited a wide range of associations (r = −0.367 to 0.944;
p < 0.05) with other nutrients in different HGEs and generated five (
X-axis) and six (
Y-axis) clusters (
p < 0.05) at the 40% linkage distance (
Figure 4B). The smallest cluster (1;
Y-axis) was characterized by negative associations between [K and Cu] with [C, N and S], while the largest (5) was comprised of 18 combinations of six nutrients and three HGEs. Most associations were positive; however, a few small negative ones were found with nutrients in the SST group (notably with K and Cu). On average, [Mg, P, Mn, S and Zn], as a group, were the most interactive (
p < 0.05) with other nutrients. They differed (
p < 0.05) from [C and Fe] which followed as a second group, [Ca and N] were intermediate, and [K and Cu], were the least interactive. Means and probability distributions of r-estimates for each HGE (
Figure 4C) suggested that NST (0.81) and NSO (0.78) means did not differ (
p > 0.05), and were larger (
p < 0.05) than SSO means (0.67), which differed (
p < 0.05) from SST (0.57). The probability distributions, especially above the mean of each HGE, differed markedly between NS and SS HGs regardless of endosperm texture.
Individual macronutrients predicted FeZn-Indices with large probability (Q
2Y; 0.50 to 0.95) at all levels of classification (
Table 5). Measured and predicted FeZn-Indexes were similar in magnitude (r > 0.97;
p < 0.0001); however, S.D. estimates were generally larger than their respective RMSE estimates. The centered and scaled PLSR coefficients suggested that most macronutrients [C, Ca, Mg, P and S] influenced the FeZn-Index positively, with a few exceptions (e.g., P in SS −0.29). Invariably, [K and N] had the largest negative coefficients for all sub-groups. Three macronutrients [P, C and S] in decreasing order, had the largest positive coefficients as compared to [Ca and Mg]. Variation (R
2X) extracted by PLSR models from independent variables ranged from 0.65 (SS) to 0.92 (OP) and accounted for 0.50 (SS) to 0.94 (OP) of Q
2Y in the FeZn-Index. However, 50% of Q
2Y estimates were >0.90 and mostly were attributed to genotypic effects, while the remaining large estimates were associated with IB (except S3). Numerical differences between Q
2Y estimates were observed between each SS (0.50) and NS (0.75), between OE (0.54) and TE (0.67) textures, and between SSO (0.50) and SST (0.54), on the one hand, and NSO (0.75) and NST (0.78), on the other. Differences between IB sub-groups were minimal, except for the smaller value (0.83) for S3. In addition, both S.D. and RMSE estimates decreased as the level of classification (i.e., sub-groups) moved from main groups (SS and NS) to genotypes.
3.5. Modeling Nutrient-Color Space Association
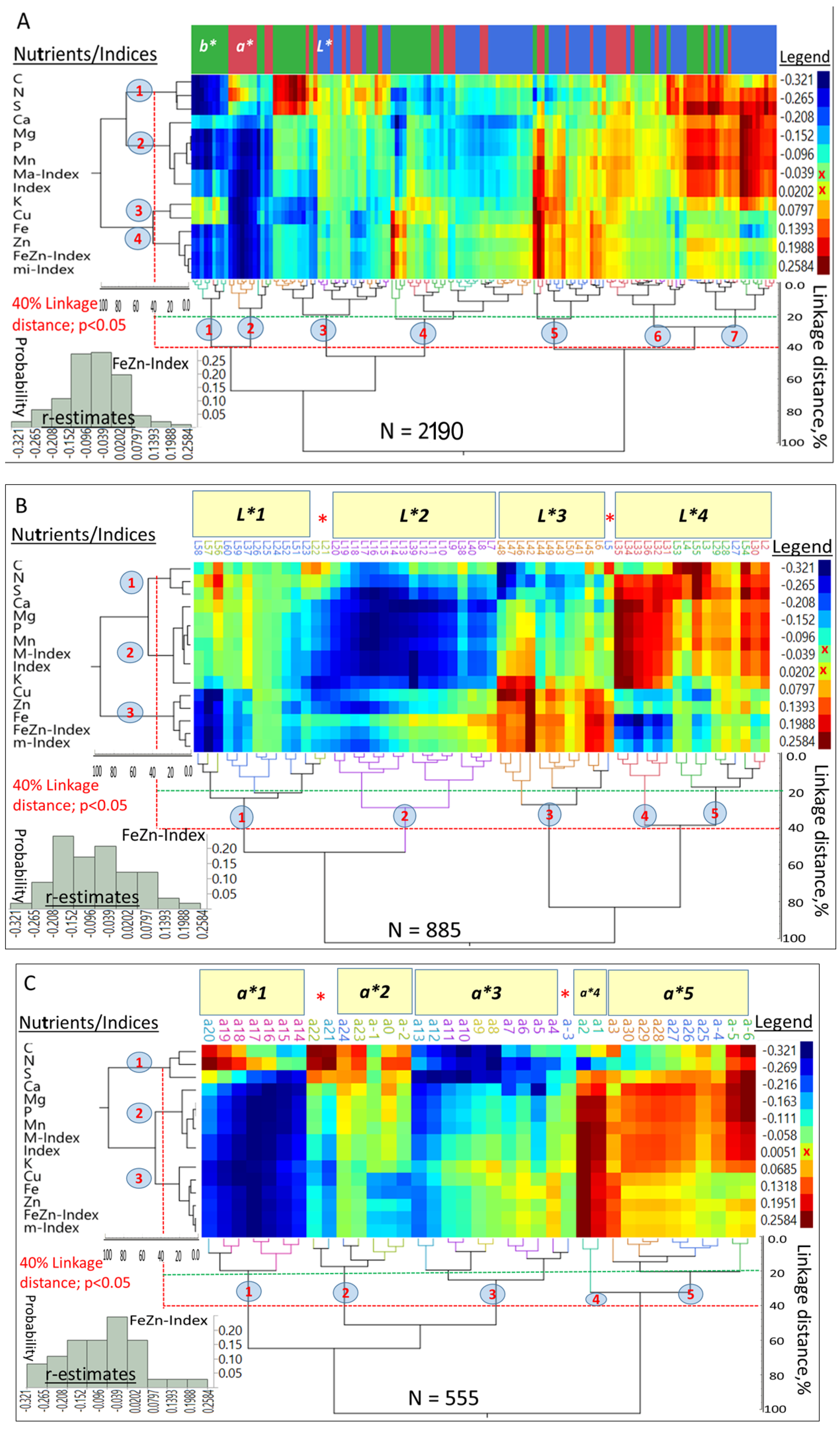
Hierarchical two-way clustering provided selection criteria based on associations between
L*a*b* and densities of 11 nutrients and their indices (
Figure 5A), as well as
L* (
Figure 5B),
a* (
Figure 5C), and
b* (
Figure 5D). At the whole color space level,
Figure 5A summarized 2190 observations, and at the 40% linkage distances, macro- and micronutrients and their respective indices (
Y-axis) were separated into four clusters (
p < 0.05), while seven clusters were formed by the color space coordinates (
X-axis). The r-estimates between both sets of variables ranged from −0.321 to 0.258.
An overall “Index” was closely associated with M-Index; the FeZn-Index was closely associated with the m-Index, while K and Cu formed an intermediate cluster between macro- and micronutrients and displayed different patterns of associations when compared with most other nutrients. Large differences in r-estimates were found between a* (−0.32 to 0.25), b* (−0.28 to 0.26), L* (−0.15 to 0.21) and different [nutrients]. Mean r-estimates between a* (−0.11), b* (−0.08), L* (−0.04) and the overall Index differed significantly from each other (p < 0.05) and suggested that they were, in decreasing order, negatively skewed. The r-estimates between the M-Index and a* (−0.25 to 0.09), b* (−0.27 to 0.08), and L* (−0.12 to 0.12) supported this suggestion. Both the m-Index and FeZn-Index showed similar trends in their r-estimates with a* (−0.22 to 0.18), b* (−0.20 to 0.15), and L* (−0.11 to 0.09).
Nutrients (and their indices) in combination with 59
L* coordinates formed five main and several minor clusters (
Figure 5B) reflecting the large variation in associations between nutrients and
L*. Nutrients and their indices formed three clusters (
Y-axis); [C, N and S] formed the first, all cations and Mn formed the second, and micronutrients formed the third cluster, at increasing linkage distances. Four L* clusters (
X-axis) summarized 885 r-estimates and demarcated some unique combinations of macro- or micronutrients.
L*1 (“L58-L21”) separated macro- from micronutrients;
L*2 captured negative associations with all nutrients;
L*3 differed from the preceding one by having relatively positive correlations with micronutrients; and
L*4 defined strong associations with macronutrients. However, two sub-clusters (4 and 5) can be identified within this cluster based on the association with macro- or micronutrients.
A small number of
a* coordinates (37) in combinations with [nutrients] formed four major clusters and one minor one (
X-axis;
Figure 5C), while [nutrients] and Indices formed three clusters (
Y-axis) similar to those in
Figure 5B. However, K clustered with micronutrients.
a*1 separated C, N, and S from other nutrients, and
a*2 featured relatively strong associations with C, N and S. However, no clear trend can be identified for association with other nutrients.
a*3 showed strong negative correlations with most macronutrients (except a-3);
a*4 had strong positive correlation of a small band (a1–a2) with most nutrients, except C and N; and
a*5 had broad positive correlations with macronutrients, but mostly small (
p > 0.05) correlations with micronutrients.
Nutrients (and their indices), in a 2-way clustering with the
b* (yellow–blue) coordinates, formed four clusters (
Figure 5D;
Y-axis) similar in their general composition to those in
Figure 5C; however, Cu and K formed a separate cluster. Six main clusters were formed by
b* (
X-axis), while two small sub-clusters (b5–b7, and b2–b3) were separated from
b*3 due to strong positive associations with most nutrients.
b*1 had widespread positive associations, especially with P and associated cations;
b*2 was dominated by negative and positive associations with macro- and micronutrients, respectively;
b*3 largely showed non-significant to slightly positive associations with macronutrient (except b5–b7, and b2–b3);
b*4 had strong negative associations with almost all nutrients (however, K and Cu were the exception); and
b*5 was vertically stratified and had strong positive associations with C, S and N, followed by largely non-significant associations with most macronutrients, and negative associations with most micronutrients, but with strong negative associations with K and Cu.
Comparisons between NS and SS, between OE and TE, and between genotypes within HGs suggested that
L*a*b* accounted for a wide range of significant (
p < 0.05) Q
2Y values (
Table 6). The largest Q
2Y estimates in NS were found for S (
b*; 0.05) and Fe (
b*; 0.07), and in SS they were found for K (
L*; 0.13; and
a*; 0.12), and Cu (
a*; 0.11). The Q
2Y estimates were generally <0.08 for OE/TE textures; the largest estimates for OE and TE were found for N (
b*; 0.08) and Mn (
b*; 0.05), respectively. Genotypes within NS exhibited small (e.g., LH82-AR16021) and large Q
2Y estimates (GQ-N16-N12). However, some genotypes within SS displayed uniform Q
2Y estimates across macro- and micro-nutrients (e.g.,
b* in LH119-AR16035), while others (e.g., DKXL370-S11) had very few and variable estimates (i.e., 0.12 and 0.05 for
a* and
b*, respectively). The Q
2Y estimates for the M-Index ranged from small (0.05) to relatively large (0.29;
L*) values, while the respective values for the m-Index were 0.05 and 0.32 (
L*). The Q
2Y estimates for the FeZn-Index were similar to those of one or both micronutrients, with exceptionally large values (e.g., DKXL370-N11a-N20 and GQ-N16-N12). Summary statistics indicated that
L*a*b* combined explained 33.3 and 25.0% of Q
2Y values in macro- and micro-nutrients in NS, respectively; the respective values for SS were 72.0 and 67.0%. Similarly, they explained 17.0% and 8.0% of Q
2Y in macro- and micro-nutrients in OE, and 28.0 and 33.0% in TE, respectively. The Q
2Y estimates for genotypes within NS were significant in 31.0 and 28.0% in macro- and micro-nutrients, respectively, while the respective values for genotypes within SS were 35.0% and 40.0%. Moreover,
L*,
a*, and
b*, accounted for variable Q
2Y estimates for macro- and micro-nutrients in NS genotypes (41.0% and 31.0%, respectively), and SS genotypes (29.0% and 25.0%, respectively). The respective values for
a* were 19.0% and 17.0% for NS genotypes, and 21.0% and 50.0% for SS genotypes, while those for
b* were 54.0% and 25.0% for NS genotypes, and 50.0 and 50.0% for SS genotypes. Significant Q
2Y estimates for the M-Index and m-Index were 44.0% and 41.0%, respectively, while 33.0% and 50.0% were the respective values for the FeZn-Index.
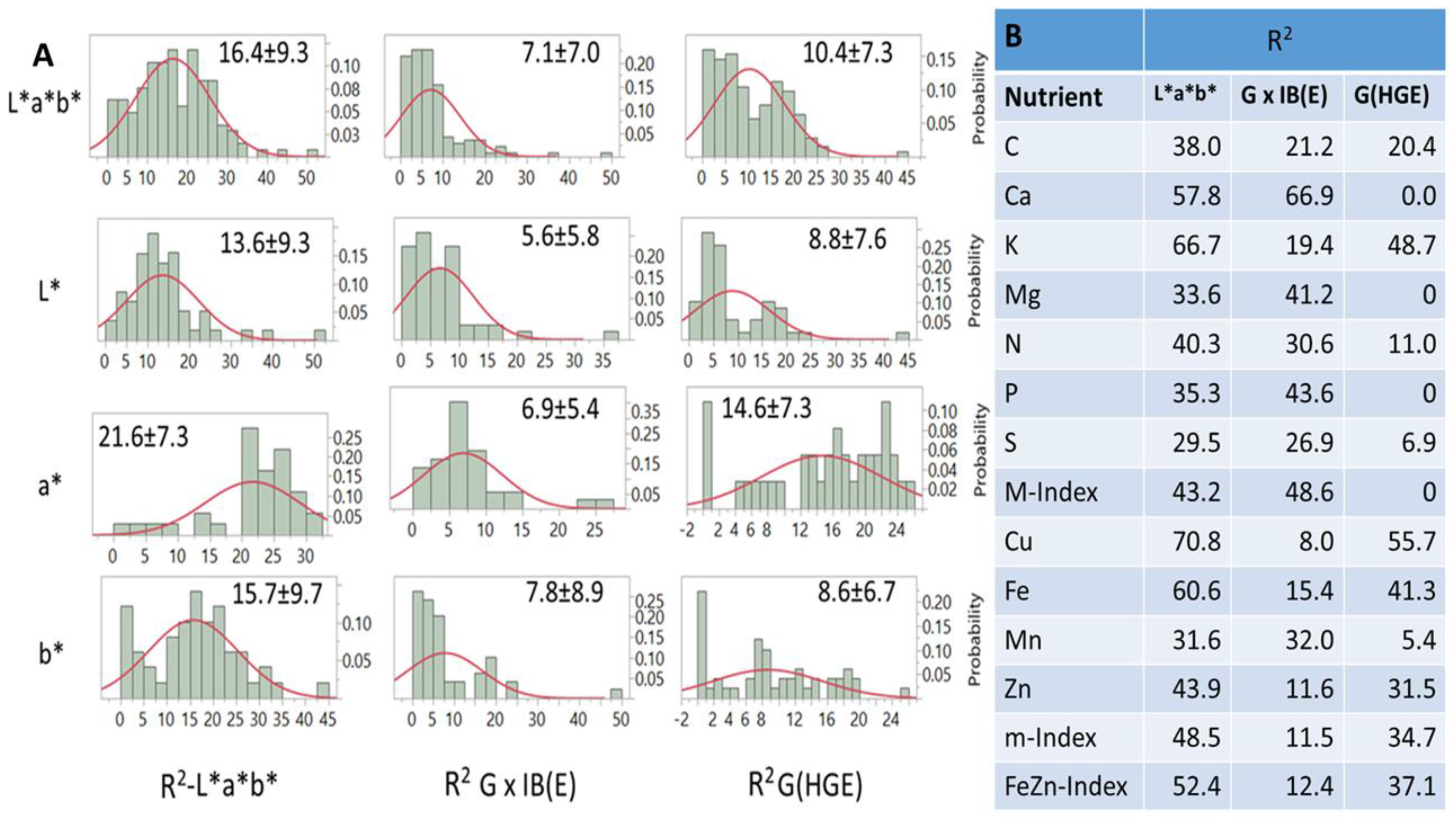
Frequency distributions of R
2 values generated by PLSR models and explained by variation in
L*a*b*, G × IB(E), and G(HGE) differed between and within factors (
Figure 6A) and between nutrients and their indices (
Figure 6B). On average, R
2 estimates ranged from zero to ~50.0, however, with mostly negatively skewed probability distributions (
Figure 6A). For all distributions, summary statistics indicated that correlations between
L*a*b* and G(HGE) and G × IB(E) were positive (r = 0.66; and 0.59;
p < 0.001), while those between the latter two were negative (r = −0.14;
p < 0.05). In addition, differences between
a* (21.6),
b* (15.7) and L* (13.6) for R
2-
L*a*b*; between
a* (14.6) and
L* (8.8) and
b* (8.6) for G(HGE); and between
b* (7.8) and
L* (6.57) and
a* (6.92) for G × IB(E) were all significant. The same R
2 estimates for each nutrient and index (
Figure 6B) averaged over macronutrients were 43.0, 35.7, and 12.4%, respectively, while the respective estimates for micronutrients were 51.7, 16.8, and 33.3%. Index values were close to those used in their estimation. Differences in variation (R
2) explained by each
L*a*b factor were found for almost all nutrients and their indices (
Figure 6B). Averaged over nutrients, *, G × IB(E), and G(HGE) explained 43.0, 35.7, and 12.4% of variation, respectively. The respective averages for micronutrients were 51.7, 16.8, and 33.5%. Some (but not all) averages were in line with R
2 estimates for the respective indices, while pair estimates for some nutrients (e.g., K/Cu; and Mg/Mn) across factors were similar.