Estimating Biomass of Black Oat Using UAV-Based RGB Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Site
2.2. UAV Platform
2.3. Data Acquisition
2.4. Data Processing and Analysis
2.5. Statistical Analysis
3. Results
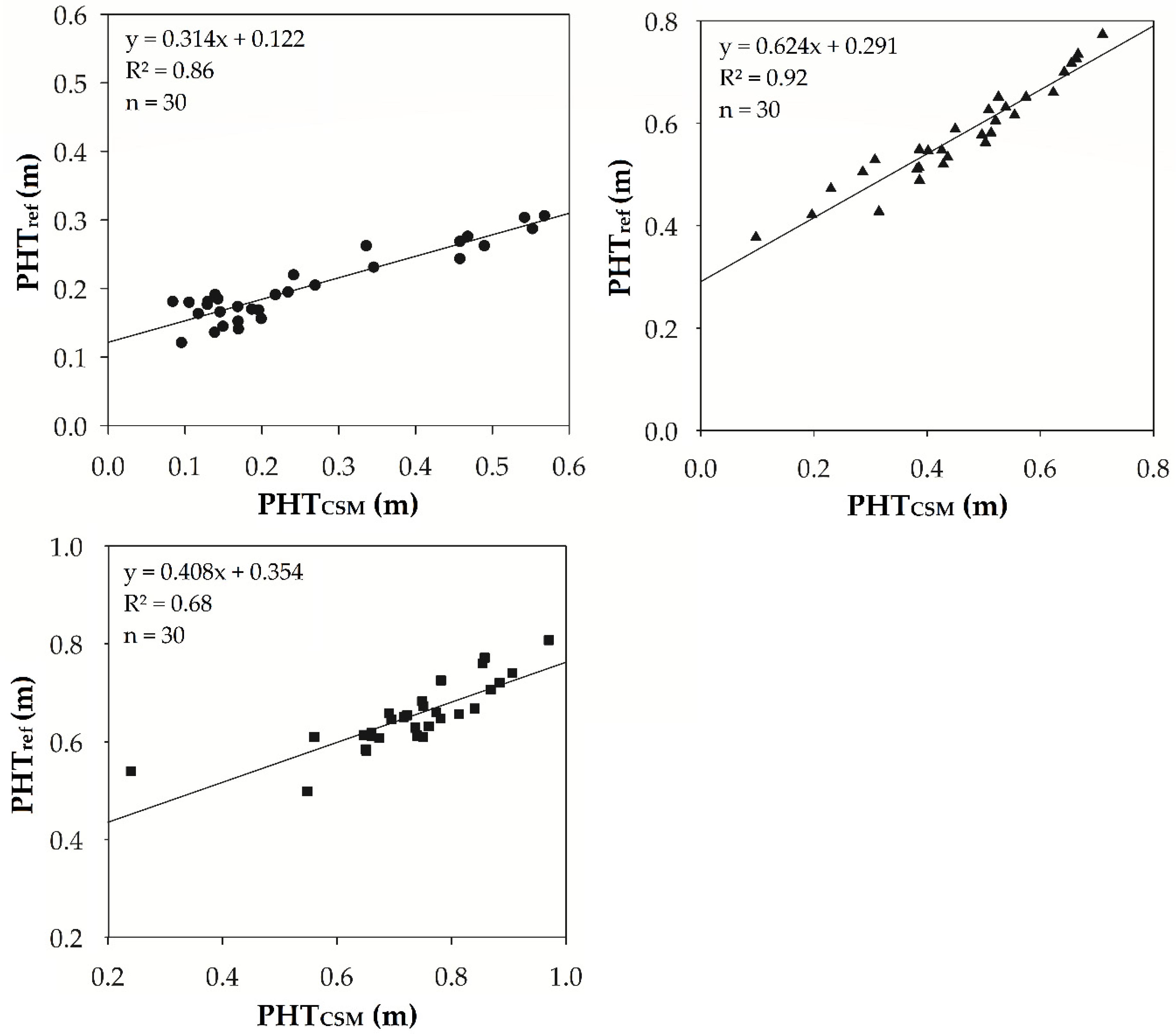
3.1. Regression Analysis
3.1.1. Plant Height
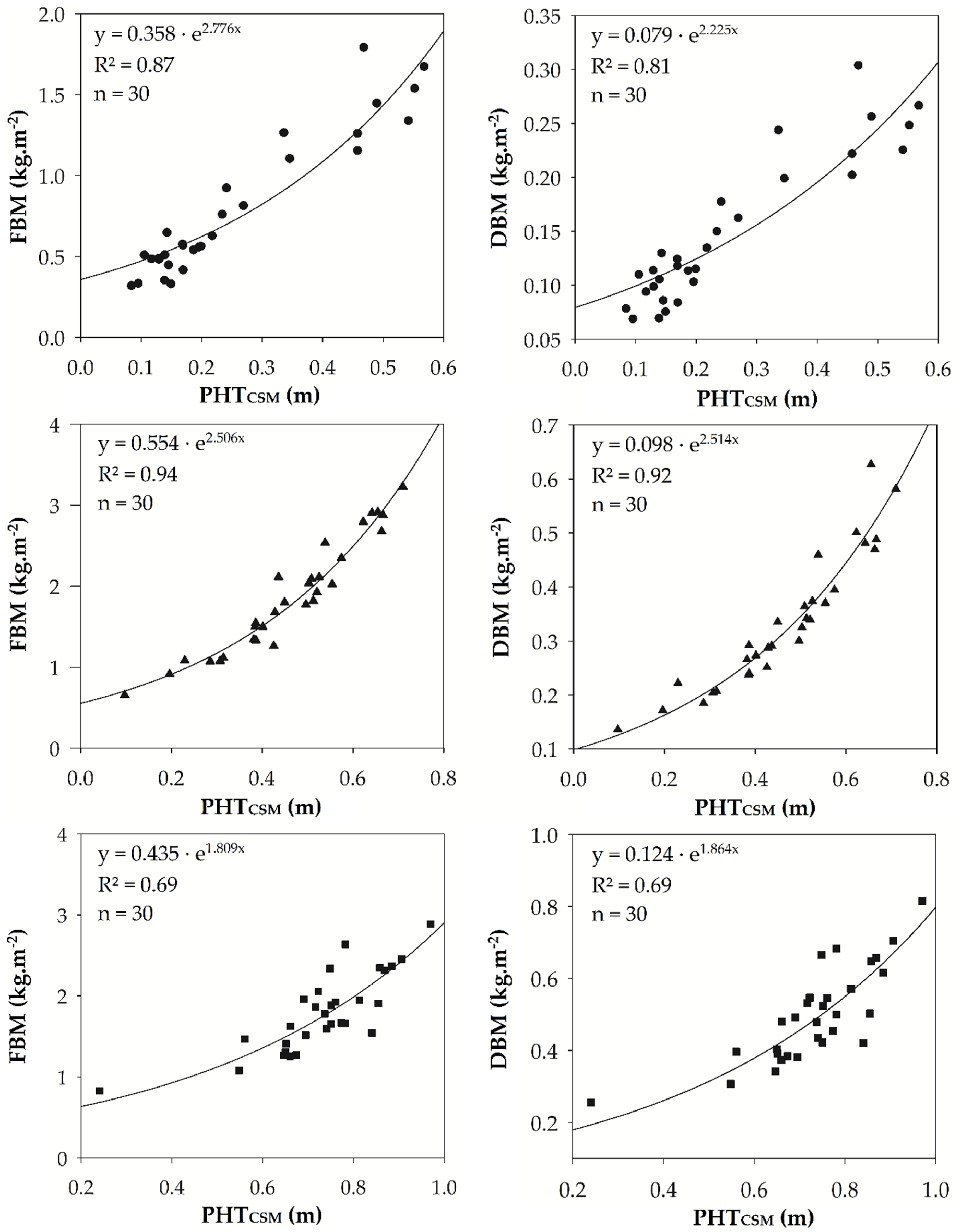
3.1.2. Biomass
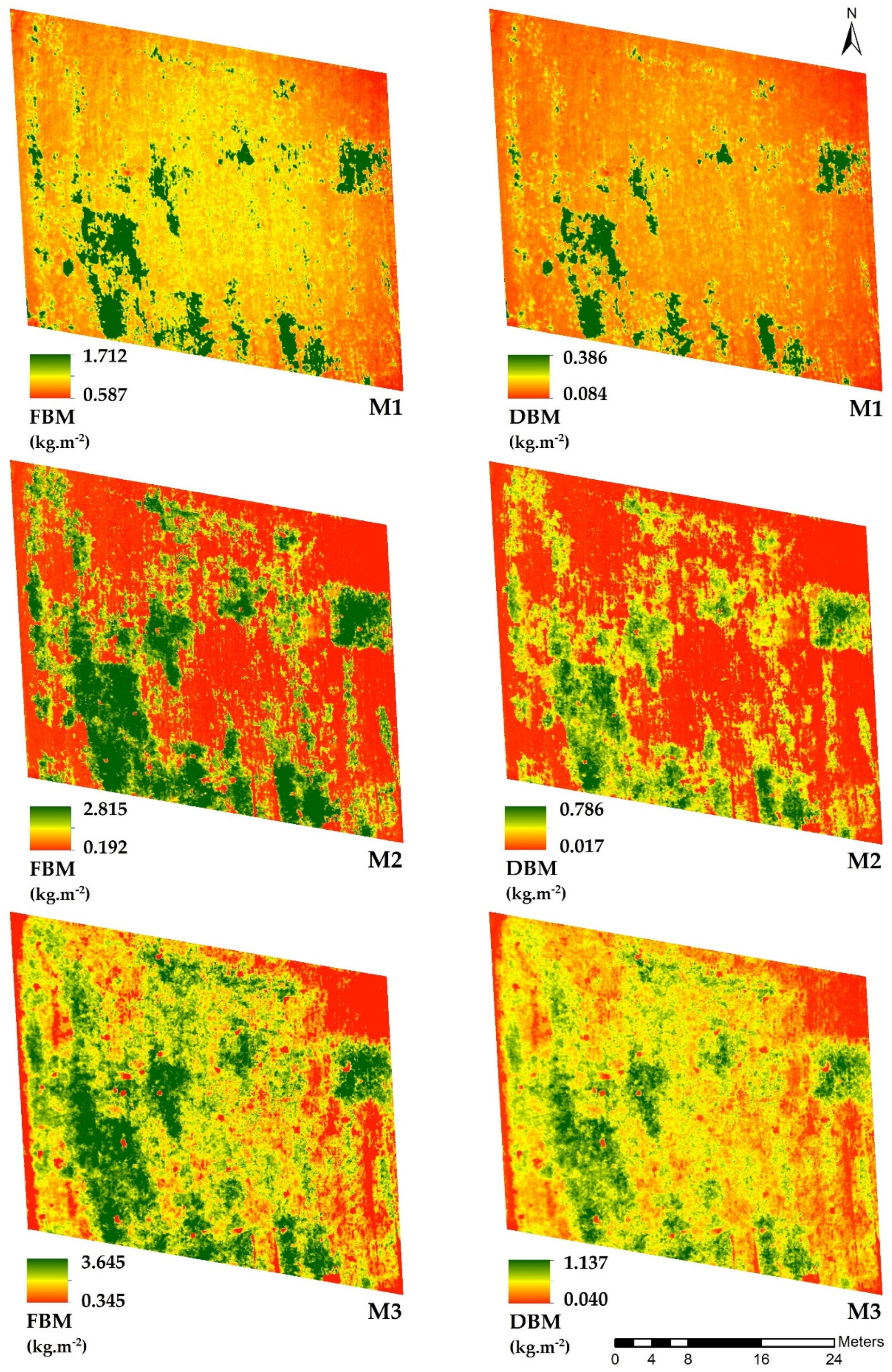
3.2. Biomass Prediction Maps
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Schirrmann, M.; Giebel, A.; Gleiniger, F.; Pflanz, M.; Lentschke, J.; Dammer, K.-H. Monitoring Agronomic Parameters of Winter Wheat Crops with Low-Cost UAV Imagery. Remote Sens. 2016, 8, 706. [Google Scholar] [CrossRef]
- Thorp, K.R.; Wang, G.; West, A.L.; Moran, M.S.; Bronson, K.F.; White, J.W.; Mon, J. Estimating crop biophysical properties from remote sensing data by inverting linked radiative transfer and ecophysiological models. Remote Sens. Environ. 2012, 124, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Niu, Z.; Huang, N.; Wang, C.; Gao, S.; Wu, C. Airborne lidar technique for estimating biomass components of maize: A case study in Zhangye City, Northwest China. Ecol. Indic. 2015, 57, 486–496. [Google Scholar] [CrossRef]
- Fischer, R.A. Irrigated spring wheat and timing and amount of nitrogen fertilizer. II. Physiology of grain yield response. Field Crops Res. 1993, 33, 57–80. [Google Scholar] [CrossRef]
- Boukerrou, L.; Rasmusson, D.D. Breeding for high biomass yield in Spring Barley. Crop Sci. 1990, 30, 31–35. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley Using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Haboudane, D.; Tremblay, N.; Wang, J.; Vigneault, P.; Li, B. New spectral indicator assessing the efficiency of crop nitrogen treatment in corn and wheat. Remote Sens. Environ. 2010, 114, 1987–1997. [Google Scholar] [CrossRef]
- Tilly, N.; Hoffmeister, D.; Cao, Q.; Huang, S.; Lenz-Wiedemann, V.I.S.; Miao, Y.; Bareth, G. Multitemporal crop surface models: Accurate plant height measurement and biomass estimation with terrestrial laser scanning in paddy rice. J. Appl. Remote Sens. 2014, 8, 83671. [Google Scholar] [CrossRef]
- Prost, L.; Jeuffroy, M.-H. Replacing the nitrogen nutrition index by the chlorophyll meter to assess wheat N status. Agron. Sust. Dev. 2007, 27, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Possoch, M.; Bieker, S.; Hoffmeister, D.; Bolten, A.; Schellberg, J.; Bareth, G. Multi-temporal crop surface models combined with the RGB vegetation index from UAV-based images for forage monitoring in grassland. Int. Arch. Photogram. Rem. Sens. Spat. Inform. Sci. 2016, XLI-B1, 991–998. [Google Scholar] [CrossRef]
- Cargnelutti Filho, A.; Toebe, M.; Alves, B.M.; Burin, C.; dos Santos, G.O.; Facco, G.; Neu, I.M.M. Linear relations among characters of black oat. Ciênc Rural 2015, 45, 985–992. [Google Scholar] [CrossRef]
- Kliemann, H.J.; Braz, A.J.P.B.; Silveira, P.M. Taxas de decomposição de resíduos de espécies de cobertura em latossolo vermelho distroférrico. Rev. Agropec. Trop. 2006, 36, 21–28. Available online: https://www.revistas.ufg.br/pat/article/view/2165/2116 (accessed on 15 January 2019).
- Calvo, C.L.; Foloni, J.S.S.; Brancalião, S.R. Produtividade de fitomassa e relação C/N de monocultivo e consórcios de guandu-anão, milheto e sorgo em três épocas de corte. Bragantia. 2010, 69, 77–86. [Google Scholar] [CrossRef]
- Lumme, J.; Karjalainen, M.; Kaartinen, H.; Kukko, A.; Hyyppä, J.; Hyyppä, H.; Jaakkola, A.; Kleemola, J. Terrestrial laser scanning of agricultural crops. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 37, 563–566. Available online: https://pdfs.semanticscholar.org/6c45/b5f5481125a311ea46a05e7acfa1de0f2cfb.pdf (accessed on 15 January 2019).
- Hakl, J.; Hrevušová, Z.; Hejcman, M.; Fuksa, P. The use of a rising plate meter to evaluate lucerne (Medicago sativa L.) height as an important agronomic trait enabling yield estimation. Grass Forage Sci. 2012, 67, 589–596. [Google Scholar] [CrossRef]
- Breda, N.J.J. Ground-based measurements of leaf area index: A review of methods, instruments and current controversies. J. Exp. Bot. 2003, 54, 2403–2417. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, T.; Tian, L.; Zhang, Y.; Ting, K.C. A review of remote sensing methods for biomass feedstock production. Biomass Bioenergy 2011, 35, 2455–2469. [Google Scholar] [CrossRef]
- Marshall, M.; Thenkabail, P. Developing in situ Non-destructive estimates of crop biomass to address issues of scale in Remote Sensing. Remote Sens. 2015, 7, 808–835. [Google Scholar] [CrossRef]
- Moeckel, T.; Safari, H.; Reddersen, B.; Fricke, T.; Wachendorf, M. Fusion of Ultrasonic and Spectral Sensor Data for Improving the Estimation of Biomass in Grasslands with Heterogeneous Sward Structure. Remote Sens. 2017, 9, 98. [Google Scholar] [CrossRef]
- Cao, Q.; Cui, Z.; Chen, X.; Khosla, R.; Dao, T.H.; Miao, Y. Quantifying spatial variability of indigenous nitrogen supply for precision nitrogen management in small scale farming. Precis. Agric. 2012, 13, 45–61. [Google Scholar] [CrossRef]
- Höfle, B. Radiometric correction of terrestrial LiDAR point cloud data for individual maize plant detection. IEEE Geosci. Remote Sens. Lett. 2014, 1, 94–98. [Google Scholar] [CrossRef]
- Bendig, J.V. Unmanned aerial vehicles (UAVs) for multi-temporal crop surface modelling—A new method for plant height and biomass estimation based on RGB-imaging. Ph.D. Thesis, University of Cologne, Cologne, Germany, 12 January 2014. Available online: https://kups.ub.uni-koeln.de/6018 (accessed on 15 January 2019).
- Berni, J.A.J.; Zarco-Tejada, P.J.; Suárez, L.; Fereres, E. Thermal and narrowband multispectral remote sensing for vegetation monitoring from an unmanned aerial vehicle. IEEE Trans. Geosci. Remote Sens. 2009, 47, 722–738. [Google Scholar] [CrossRef]
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Swain, K.C.; Zaman, Q.U. Rice crop monitoring with unmanned helicopter remote sensing images. In Remote Sensing of Biomass—Principles and Applications; Fatoyinbo, L., Ed.; INTECH Open Access Publisher: Rijeka, Croatia, 2012; pp. 253–272. [Google Scholar]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Verhoeven, G. Taking computer vision aloft—Archaeological three-dimensional reconstructions from aerial photographs with photoscan. Archaeol. Prospect. 2011, 18, 67–73. [Google Scholar] [CrossRef]
- Hoffmeister, D.; Bolten, A.; Curdt, C.; Waldhoff, G.; Bareth, G. High resolution Crop Surface Models (CSM) and Crop Volume Models (CVM) on field level by terrestrial laser scanning. Proc. SPIE 2010, 7840. [Google Scholar] [CrossRef]
- Li, W.; Niu, Z.; Chen, H.; Li, D.; Wu, M.; Zhao, W. Remote estimation of canopy height and aboveground biomass of maize using high-resolution stereo images from a low-cost unmanned aerial vehicle system. Ecol. Indic. 2016, 67, 637–648. [Google Scholar] [CrossRef]
- Ballesteros, R.; Ortega, J.F.; Hernandez, D.; Moreno, M.A. Onion biomass monitoring using UAV-based RGB imaging. Precis. Agric. 2018, 19, 840–857. [Google Scholar] [CrossRef]
- Grüner, E.; Astor, T.; Wachendorf, M. Biomass Prediction of Heterogeneous Temperate Grasslands Using an SfM Approach Based on UAV Imaging. Agronomy 2019, 9, 54. [Google Scholar] [CrossRef]
- Cooper, S.; Roy, D.; Schaaf, C.; Paynter, I. Examination of the Potential of Terrestrial Laser Scanning and Structure-from-Motion Photogrammetry for Rapid Nondestructive Field Measurement of Grass Biomass. Remote Sens. 2017, 9, 531. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Scotford, I.M.; Miller, P.C.H. Combination of spectral reflectance and ultrasonic sensing to monitor the growth of winter wheat. Biosyst. Eng. 2004, 87, 27–38. [Google Scholar] [CrossRef]
- Roth, L.; Streit, B. Predicting cover crop biomass by lightweight UAS-based RGB and NIR photography: An applied photogrammetric approach. Precis. Agric. 2018, 19, 93–114. [Google Scholar] [CrossRef]
- Tilly, N.; Aasen, H.; Bareth, G. Fusion of plant height and vegetation indices for the estimation of barley biomass. Remote Sens. 2015, 7, 11449–11480. [Google Scholar] [CrossRef]
- Ehlert, D.; Horn, H.-J.; Adamek, R. Measuring crop biomass density by laser triangulation. Comput. Electron. Agric. 2008, 61, 117–125. [Google Scholar] [CrossRef]
- Eitel, J.U.H.; Magney, T.S.; Vierling, L.A.; Brown, T.T.; Huggins, D.R. LiDAR based biomass and crop nitrogen estimates for rapid, non-destructive assessment of wheat nitrogen status. Field Crop Res. 2014, 159, 21–32. [Google Scholar] [CrossRef]
- Ehlert, D.; Heisig, M.; Adamek, R. Suitability of a laser rangefinder to characterize winter wheat. Precis. Agric. 2010, 11, 650–663. [Google Scholar] [CrossRef]
- Harmoney, K.R.; Moore, K.J.; George, J.R.; Brummer, E.C.; Russell, J.R. Determination of pasture biomass using four indirect methods. Agron. J. 1997, 89, 665–672. [Google Scholar] [CrossRef]
- Erdle, K.; Mistele, B.; Schmidhalter, U. Comparison of active and passive spectral sensors in discriminating biomass parameters and nitrogen status in wheat cultivars. Field Crop Res. 2011, 124, 74–84. [Google Scholar] [CrossRef]
- Khosla, R.; Westfall, D.G.; Reich, R.M.; Mahal, J.S.; Gangloff, W.J. Spatial Variation and Site-Specific Management Zones. In Geostatistical Applications for Precision Agriculture; Oliver, M.A., Ed.; Springer Science: Berlin, Germany, 2010; pp. 195–219. [Google Scholar]


| ID | Date | Mission Objective | Growth Stage (DAS) | Wind Speed 1 | Images Collected | Point Density (pt·m−2) | Image Overlap 2 | GSD (cm·px−1) |
|---|---|---|---|---|---|---|---|---|
| M0 | 4 June 2017 | DTM | - | 1.1 | 150 | 3228 | >9 | 1.76 |
| M1 | 11 August 2017 | DSM | Booting (53) | 4.1 | 142 | 2916 | >9 | 1.85 |
| M2 | 25 August 2017 | DSM | Flowering (67) | 3.4 | 179 | 4056 | >9 | 1.57 |
| M3 | 6 September 2017 | DSM | Grain filling (79) | 3.3 | 153 | 3052 | >9 | 1.81 |
| Variable | n | Unit | Abbreviation | Mean | Min | Max | SD | Median | |
|---|---|---|---|---|---|---|---|---|---|
| Mission 1 | Reference plant height | 30 | m | PHTref | 0.20 | 0.12 | 0.31 | 0.05 | 0.18 |
| CSM plant height | 30 | m | PHTCSM | 0.25 | 0.08 | 0.57 | 0.15 | 0.19 | |
| Fresh Biomass | 30 | kg·m−2 | FBM | 0.795 | 0.320 | 1.793 | 0.440 | 0.573 | |
| Dry Biomass | 30 | kg·m−2 | DBM | 0.149 | 0.069 | 0.304 | 0.068 | 0.121 | |
| Mission 2 | Reference plant height | 30 | m | PHTref | 0.58 | 0.38 | 0.77 | 0.10 | 0.57 |
| CSM plant height | 30 | m | PHTCSM | 0.46 | 0.10 | 0.71 | 0.15 | 0.47 | |
| Fresh Biomass | 30 | kg·m−2 | FBM | 1.869 | 0.655 | 3.230 | 0.685 | 1.811 | |
| Dry Biomass | 30 | kg·m−2 | DBM | 0.334 | 0.136 | 0.627 | 0.123 | 0.313 | |
| Mission 3 | Reference plant height | 30 | m | PHTref | 0.65 | 0.50 | 0.81 | 0.07 | 0.65 |
| CSM plant height | 30 | m | PHTCSM | 0.73 | 0.24 | 0.97 | 0.14 | 0.74 | |
| Fresh Biomass | 30 | kg·m−2 | FBM | 1.794 | 0.829 | 2.885 | 0.482 | 1.723 | |
| Dry Biomass | 30 | kg·m−2 | DBM | 0.497 | 0.255 | 0.815 | 0.130 | 0.486 |
| Y | x | Model | n | R2 | ME | MAE | RMSE | rRMSE | |
|---|---|---|---|---|---|---|---|---|---|
| Mission 1 | PHTref | PHTCSM | 0.314x + 0.122 | 30 | 0.86 | 0.000 | 0.016 | 0.019 | 9.6 |
| FBM | PHTCSM | 0.358 · exp(2.776x) | 30 | 0.87 | 0.005 | 0.111 | 0.155 | 19.5 | |
| DBM | PHTCSM | 0.079 · exp(2.255x) | 30 | 0.81 | 0.000 | 0.022 | 0.029 | 19.3 | |
| Mission 2 | PHTref | PHTCSM | 0.624 x + 0.291 | 30 | 0.92 | 0.000 | 0.024 | 0.028 | 4.8 |
| FBM | PHTCSM | 0.554 · exp(2.506x) | 30 | 0.94 | 0.003 | 0.126 | 0.165 | 8.9 | |
| DBM | PHTCSM | 0.098 · exp(2.514x) | 30 | 0.92 | -0.002 | 0.025 | 0.035 | 10.5 | |
| Mission 3 | PHTref | PHTCSM | 0.408x + 0.354 | 30 | 0.68 | 0.000 | 0.030 | 0.037 | 5.7 |
| FBM | PHTCSM | 0.435 · exp(1.899x) | 30 | 0.69 | 0.001 | 0.208 | 0.265 | 14.8 | |
| DBM | PHTCSM | 0.124 · exp(1.864x) | 30 | 0.69 | 0.001 | 0.058 | 0.071 | 14.3 |
| y | x | Model | n | R² | ME | MAE | RMSE | rRMSE | |
|---|---|---|---|---|---|---|---|---|---|
| Cal. | FBM | PHTCSM | 0.707 · exp(1.438x) | 63 | 0.64 | 0.013 | 0.354 | 0.438 | 29.1 |
| DBM | PHTCSM | 0.109 · exp(2.056x) | 63 | 0.89 | 0.004 | 0.048 | 0.064 | 19.1 | |
| Val. | FBM | PHTCSM | 1.206x − 0.3220 | 27 | 0.52 | 0.020 | 0.394 | 0.506 | 35.2 |
| DBM | PHTCSM | 1.086x − 0.0335 | 27 | 0.84 | 0.006 | 0.052 | 0.063 | 20.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acorsi, M.G.; das Dores Abati Miranda, F.; Martello, M.; Smaniotto, D.A.; Sartor, L.R. Estimating Biomass of Black Oat Using UAV-Based RGB Imaging. Agronomy 2019, 9, 344. https://doi.org/10.3390/agronomy9070344
Acorsi MG, das Dores Abati Miranda F, Martello M, Smaniotto DA, Sartor LR. Estimating Biomass of Black Oat Using UAV-Based RGB Imaging. Agronomy. 2019; 9(7):344. https://doi.org/10.3390/agronomy9070344
Chicago/Turabian StyleAcorsi, Matheus Gabriel, Fabiani das Dores Abati Miranda, Maurício Martello, Danrley Antonio Smaniotto, and Laercio Ricardo Sartor. 2019. "Estimating Biomass of Black Oat Using UAV-Based RGB Imaging" Agronomy 9, no. 7: 344. https://doi.org/10.3390/agronomy9070344





