14-3-3 Proteins are Regulators of Autophagy
Abstract
:1. Introduction
1.1. Overview of 14-3-3 Proteins
1.2. Role of 14-3-3 in Apoptosis
1.3. Autophagy Process Summary
2. Role of 14-3-3 Proteins in Autophagy
2.1. Regulation of TCS2 by 14-3-3 Proteins
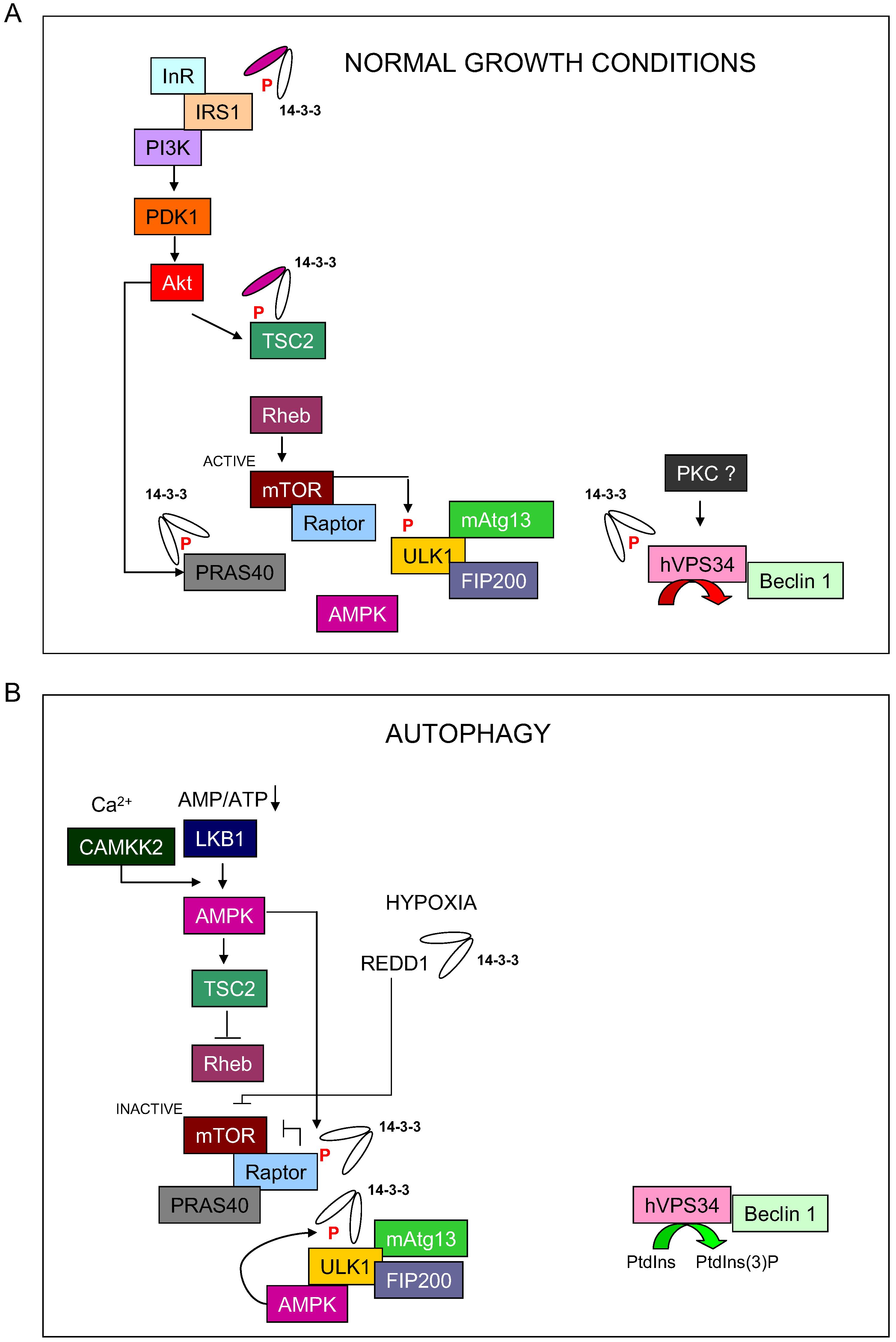
2.2. Regulation of PRAS40 by 14-3-3 Proteins
2.3. Regulation of Raptor by 14-3-3 Proteins
2.4. Regulation of REDD by 14-3-3 Proteins
2.5. 14-3-3 Proteins Regulate MAPK Pathway
2.6. 14-3-3 Proteins Regulate Protein Kinase C
2.7. 14-3-3 Proteins Regulate CaMKKβ
3. Role of 14-3-3 Proteins during Autophagy Initiation Process
3.1. 14-3-3 Proteins Regulate ULK1
3.2. 14-3-3 Proteins Regulate hVps34 (PI3KC3)
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Chaudhri, M.; Scarabel, M.; Aitken, A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 2003, 300, 679–685. [Google Scholar] [CrossRef]
- Jones, D.H.; Ley, S.; Aitken, A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995, 368, 55–58. [Google Scholar] [CrossRef]
- Mackintosh, C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004, 381, 329–342. [Google Scholar] [CrossRef]
- Muslin, A.J.; Tanner, J.W.; Allen, P.M.; Shaw, A.S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996, 84, 889–897. [Google Scholar] [CrossRef]
- Yaffe, M.B.; Rittinger, K.; Volinia, S.; Caron, P.R.; Aitken, A.; Leffers, H.; Gamblin, S.J.; Smerdon, S.J.; Cantley, L.C. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 1997, 91, 961–971. [Google Scholar] [CrossRef]
- Johnson, C.; Crowther, S.; Stafford, M.J.; Campbell, D.G.; Toth, R.; MacKintosh, C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010, 427, 69–78. [Google Scholar] [CrossRef]
- Aitken, A.; Howell, S.; Jones, D.; Madrazo, J.; Patel, Y. 14-3-3 alpha and delta are the phosphorylated forms of raf-activating 14-3-3 beta and zeta. In vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys MOTIF. In J. Biol. Chem.; 1995; Volume 270, pp. 5706–5709. [Google Scholar]
- Moreira, J.M.; Shen, T.; Ohlsson, G.; Gromov, P.; Gromova, I.; Celis, J.E. A combined proteome and ultrastructural localization analysis of 14-3-3 proteins in transformed human amnion (AMA) cells: definition of a framework to study isoform-specific differences. Mol. Cell. Proteomics 2008, 7, 1225–1240. [Google Scholar] [CrossRef]
- Kilani, R.T.; Medina, A.; Aitken, A.; Jalili, R.B.; Carr, M.; Ghahary, A. Identification of different isoforms of 14-3-3 protein family in human dermal and epidermal layers. Mol. Cell. Biochem. 2008, 314, 161–169. [Google Scholar] [CrossRef]
- Yaffe, M.B. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002, 513, 53–57. [Google Scholar] [CrossRef]
- Wilker, E.; Yaffe, M.B. 14-3-3 Proteins—a focus on cancer and human disease. J. Mol. Cell. Cardiol. 2004, 37, 633–642. [Google Scholar] [CrossRef]
- Meek, S.E.; Lane, W.S.; Piwnica-Worms, H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 2004, 279, 32046–32054. [Google Scholar] [CrossRef]
- Pozuelo Rubio, M.; Geraghty, K.M.; Wong, B.H.; Wood, N.T.; Campbell, D.G.; Morrice, N.; Mackintosh, C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004, 379, 395–408. [Google Scholar] [CrossRef]
- Jin, J.; Smith, F.D.; Stark, C.; Wells, C.D.; Fawcett, J.P.; Kulkarni, S.; Metalnikov, P.; O’Donnell, P.; Taylor, P.; Taylor, L.; Zougman, A.; Woodgett, J.R.; Langeberg, L.K.; Scott, J.D.; Pawson, T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004, 14, 1436–1450. [Google Scholar] [CrossRef]
- Kjarland, E.; Keen, T.J.; Kleppe, R. Does isoform diversity explain functional differences in the 14-3-3 protein family’? Curr. Pharm. Biotechnol. 2006, 7, 217–223. [Google Scholar] [CrossRef]
- Pozuelo-Rubio, M. Proteomic and biochemical analysis of 14-3-3-binding proteins during C2- ceramide-induced apoptosis. FEBS J. 2010, 277, 3321–3342. [Google Scholar] [CrossRef]
- Chang, I.F.; Curran, A.; Woolsey, R.; Quilici, D.; Cushman, J.C.; Mittler, R.; Harmon, A.; Harper, J.F. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009, 9, 2967–2985. [Google Scholar] [CrossRef]
- Puri, P.; Myers, K.; Kline, D.; Vijayaraghavan, S. Proteomic analysis of bovine sperm YWHA binding partners identify proteins involved in signaling and metabolism. Biol. Reprod. 2008, 79, 1183–1191. [Google Scholar] [CrossRef]
- Benzinger, A.; Muster, N.; Koch, H.B.; Yates, J.R., 3rd; Hermeking, H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol. Cell. Proteomics 2005, 4, 785–795. [Google Scholar] [CrossRef]
- Masters, S.C.; Subramanian, R.R.; Truong, A.; Yang, H.; Fujii, K.; Zhang, H.; Fu, H. Survival-promoting functions of 14-3-3 proteins. Biochem. Soc. Trans. 2002, 30, 360–365. [Google Scholar]
- Zha, J.; Harada, H.; Yang, E.; Jockel, J.; Korsmeyer, S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 1996, 87, 619–628. [Google Scholar] [CrossRef]
- Nomura, M.; Shimizu, S.; Sugiyama, T.; Narita, M.; Ito, T.; Matsuda, H.; Tsujimoto, Y. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J. Biol. Chem. 2003, 278, 2058–2065. [Google Scholar]
- Zhang, L.; Chen, J.; Fu, H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 8511–8515. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Masters, S.C.; Fu, H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001, 276, 45193–45200. [Google Scholar] [CrossRef]
- Cao, W.; Yang, X.; Zhou, J.; Teng, Z.; Cao, L.; Zhang, X.; Fei, Z. Targeting 14-3-3 protein, difopein induces apoptosis of human glioma cells and suppresses tumour growth in mice. Apoptosis 2010, 15, 230–241. [Google Scholar] [CrossRef]
- Qi, W.; Martinez, J.D. Reduction of 14-3-3 proteins correlates with increased sensitivity to killing of human lung cancer cells by ionizing radiation. Radiat. Res. 2003, 160, 217–223. [Google Scholar] [CrossRef]
- Niemantsverdriet, M.; Wagner, K.; Visser, M.; Backendorf, C. Cellular functions of 14-3-3 zeta in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene 2008, 27, 1315–1319. [Google Scholar] [CrossRef]
- Neal, C.L.; Yao, J.; Yang, W.; Zhou, X.; Nguyen, N.T.; Lu, J.; Danes, C.G.; Guo, H.; Lan, K.H.; Ensor, J.; Hittelman, W.; Hung, M.C.; Yu, D. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009, 69, 3425–3432. [Google Scholar] [CrossRef]
- Kumar, A.P.; Garcia, G.E.; Orsborn, J.; Levin, V.A.; Slaga, T.J. 2-Methoxyestradiol interferes with NF kappa B transcriptional activity in primitive neuroectodermal brain tumors: implications for management. Carcinogenesis 2003, 24, 209–216. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: a double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar] [CrossRef]
- Galluzzi, L.; Aaronson, S.A.; Abrams, J.; Alnemri, E.S.; Andrews, D.W.; Baehrecke, E.H.; Bazan, N.G.; Blagosklonny, M.V.; Blomgren, K.; Borner, C.; Bredesen, D.E.; Brenner, C.; Castedo, M.; Cidlowski, J.A.; Ciechanover, A.; Cohen, G.M.; De Laurenzi, V.; De Maria, R.; Deshmukh, M.; Dynlacht, B.D.; El-Deiry, W.S.; Flavell, R.A.; Fulda, S.; Garrido, C.; Golstein, P.; Gougeon, M.L.; Green, D.R.; Gronemeyer, H.; Hajnoczky, G.; Hardwick, J.M.; Hengartner, M.O.; Ichijo, H.; Jaattela, M.; Kepp, O.; Kimchi, A.; Klionsky, D.J.; Knight, R.A.; Kornbluth, S.; Kumar, S.; Levine, B.; Lipton, S.A.; Lugli, E.; Madeo, F.; Malomi, W.; Marine, J.C.; Martin, S.J.; Medema, J.P.; Mehlen, P.; Melino, G.; Moll, U.M.; Morselli, E.; Nagata, S.; Nicholson, D.W.; Nicotera, P.; Nunez, G.; Oren, M.; Penninger, J.; Pervaiz, S.; Peter, M.E.; Piacentini, M.; Prehn, J.H.; Puthalakath, H.; Rabinovich, G.A.; Rizzuto, R.; Rodrigues, C.M.; Rubinsztein, D.C.; Rudel, T.; Scorrano, L.; Simon, H.U.; Steller, H.; Tschopp, J.; Tsujimoto, Y.; Vandenabeele, P.; Vitale, I.; Vousden, K.H.; Youle, R.J.; Yuan, J.; Zhivotovsky, B.; Kroemer, G. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009, 16, 1093–1107. [Google Scholar]
- Kroemer, G.; Jaattela, M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer 2005, 5, 886–897. [Google Scholar] [CrossRef]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007, 8, 741–752. [Google Scholar] [CrossRef]
- Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Juhasz, G.; Goraksha-Hicks, P.; Arsham, A.M.; Mallin, D.R.; Muller, L.K.; Neufeld, T.P. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem. Soc. Trans. 2009, 37, 232–236. [Google Scholar] [CrossRef]
- Bjorkoy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar]
- Levine, B.; Yuan, J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005, 115, 2679–2688. [Google Scholar] [CrossRef]
- Baehrecke, E.H. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef]
- Gozuacik, D.; Kimchi, A. Autophagy and cell death. Curr. Top. Dev. Biol. 2007, 78, 217–245. [Google Scholar] [CrossRef]
- Lockshin, R.A.; Zakeri, Z. Programmed cell death and apoptosis: origins of the theory. Nat Rev. Mol. Cell Biol. 2001, 2, 545–550. [Google Scholar] [CrossRef]
- Daido, S.; Kanzawa, T.; Yamamoto, A.; Takeuchi, H.; Kondo, Y.; Kondo, S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004, 64, 4286–4293. [Google Scholar] [CrossRef]
- Steinberg, G.R.; Kemp, B.E. AMPK in Health and Disease. Physiol. Rev. 2009, 89, 1025–1078. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Inoki, K.; Zhu, T.; Guan, K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Corradetti, M.N.; Inoki, K.; Bardeesy, N.; DePinho, R.A.; Guan, K.L. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004, 18, 1533–1538. [Google Scholar] [CrossRef]
- Shaw, R.J.; Bardeesy, N.; Manning, B.D.; Lopez, L.; Kosmatka, M.; DePinho, R.A.; Cantley, L.C. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004, 6, 91–99. [Google Scholar] [CrossRef]
- Liu, L.; Cash, T.P.; Jones, R.G.; Keith, B.; Thompson, C.B.; Simon, M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 2006, 21, 521–531. [Google Scholar] [CrossRef]
- Shaw, R.J. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009, 196, 65–80. [Google Scholar] [CrossRef]
- Miyazaki, M.; McCarthy, J.J.; Esser, K.A. Insulin like growth factor-1-induced phosphorylation and altered distribution of tuberous sclerosis complex (TSC)1/TSC2 in C2C12 myotubes. FEBS J. 2010, 277, 2180–2191. [Google Scholar] [CrossRef]
- Cai, S.L.; Tee, A.R.; Short, J.D.; Bergeron, J.M.; Kim, J.; Shen, J.; Guo, R.; Johnson, C.L.; Kiguchi, K.; Walker, C.L. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 2006, 173, 279–289. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 2009, 37, 217–222. [Google Scholar] [CrossRef]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar] [CrossRef]
- Vander Haar, E.; Lee, S.I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef]
- Wang, L.; Harris, T.E.; Lawrence, J.C., Jr. Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J. Biol. Chem. 2008, 283, 15619–15627. [Google Scholar]
- Nascimento, E.B.; Snel, M.; Guigas, B.; van der Zon, G.C.; Kriek, J.; Maassen, J.A.; Jazet, I.M.; Diamant, M.; Ouwens, D.M. Phosphorylation of PRAS40 on Thr246 by PKB/AKT facilitates efficient phosphorylation of Ser183 by mTORC1. Cell Signal. 2010, 22, 961–967. [Google Scholar] [CrossRef]
- Fonseca, B.D.; Smith, E.M.; Lee, V.H.; MacKintosh, C.; Proud, C.G. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J. Biol. Chem. 2007, 282, 24514–24524. [Google Scholar]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Brugarolas, J.; Lei, K.; Hurley, R.L.; Manning, B.D.; Reiling, J.H.; Hafen, E.; Witters, L.A.; Ellisen, L.W.; Kaelin, W.G., Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004, 18, 2893–2904. [Google Scholar] [CrossRef] [Green Version]
- Corradetti, M.N.; Inoki, K.; Guan, K.L. The stress-induced proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2005, 280, 9769–9772. [Google Scholar]
- Sofer, A.; Lei, K.; Johannessen, C.M.; Ellisen, L.W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell Biol. 2005, 25, 5834–5845. [Google Scholar] [CrossRef]
- DeYoung, M.P.; Horak, P.; Sofer, A.; Sgroi, D.; Ellisen, L.W. Hypoxia regulates TSC1/2- mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008, 22, 239–251. [Google Scholar] [CrossRef]
- Xiang, X.; Yuan, M.; Song, Y.; Ruderman, N.; Wen, R.; Luo, Z. 14-3-3 facilitates insulin-stimulated intracellular trafficking of insulin receptor substrate 1. Mol. Endocrinol. 2002, 16, 552–562. [Google Scholar] [CrossRef]
- McKay, M.M.; Morrison, D.K. Integrating signals from RTKs to ERK/MAPK. Oncogene 2007, 26, 3113–3121. [Google Scholar] [CrossRef]
- Murphy, L.O.; Blenis, J. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 2006, 31, 268–275. [Google Scholar] [CrossRef]
- Cagnol, S.; Chambard, J.C. ERK and cell death: mechanisms of ERK-induced cell death—apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef]
- Ellington, A.A.; Berhow, M.A.; Singletary, K.W. Inhibition of Akt signaling and enhanced ERK1/2 activity are involved in induction of macroautophagy by triterpenoid B-group soyasaponins in colon cancer cells. Carcinogenesis 2006, 27, 298–306. [Google Scholar] [CrossRef]
- Choi, C.H.; Jung, Y.K.; Oh, S.H. Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol. Pharmacol. 2010, 78, 114–125. [Google Scholar] [CrossRef]
- Wang, S.H.; Shih, Y.L.; Lee, C.C.; Chen, W.L.; Lin, C.J.; Lin, Y.S.; Wu, K.H.; Shih, C.M. The role of endoplasmic reticulum in cadmium-induced mesangial cell apoptosis. Chem. Biol. Interact. 2009, 181, 45–51. [Google Scholar] [CrossRef]
- Yang, L.Y.; Wu, K.H.; Chiu, W.T.; Wang, S.H.; Shih, C.M. The cadmium-induced death of mesangial cells results in nephrotoxicity. Autophagy 2009, 5, 571–572. [Google Scholar] [CrossRef]
- Cheng, Y.; Qiu, F.; Tashiro, S.; Onodera, S.; Ikejima, T. ERK and JNK mediate TNFalpha- induced p53 activation in apoptotic and autophagic L929 cell death. Biochem. Biophys. Res.Commun. 2008, 376, 483–488. [Google Scholar] [CrossRef]
- Wang, J.; Whiteman, M.W.; Lian, H.; Wang, G.; Singh, A.; Huang, D.; Denmark, T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol.Chem. 2009, 284, 21412–21424. [Google Scholar] [CrossRef]
- Corcelle, E.; Nebout, M.; Bekri, S.; Gauthier, N.; Hofman, P.; Poujeol, P.; Fenichel, P.; Mograbi, B. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006, 66, 6861–6870. [Google Scholar]
- Tzivion, G.; Luo, Z.; Avruch, J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 1998, 394, 88–92. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Meikle, S.; Peyssonnaux, C.; Grindlay, J.; Kaiser, C.; Steen, H.; Shaw, P.E.; Mischak, H.; Eychene, A.; Kolch, W. A Raf-1 mutant that dissociates MEK/extracellular signal-regulated kinase activation from malignant transformation and differentiation but not proliferation. Mol. Cell Biol. 2003, 23, 1983–1993. [Google Scholar]
- Dumaz, N.; Marais, R. Protein kinase A blocks Raf-1 activity by stimulating 14-3-3 binding and blocking Raf-1 interaction with Ras. J. Biol. Chem. 2003, 278, 29819–29823. [Google Scholar] [CrossRef]
- Wagner, E.F.; Nebreda, A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 2009, 9, 537–549. [Google Scholar] [CrossRef]
- Webber, J.L.; Tooze, S.A. New insights into the function of Atg9. FEBS Lett. 2010, 584, 1319–1326. [Google Scholar] [CrossRef]
- Cui, Q.; Tashiro, S.; Onodera, S.; Minami, M.; Ikejima, T. Oridonin induced autophagy in human cervical carcinoma HeLa cells through Ras, JNK, and P38 regulatio. J. Pharmacol. Sci. 2007, 105, 317–325. [Google Scholar] [CrossRef]
- Liao, P.C.; Ng, L.T.; Lin, L.T.; Richardson, C.D.; Wang, G.H.; Lin, C.C. Resveratrolarrests cell cycle and induces apoptosis in human hepatocellular carcinoma Huh-7 cells. J. Med. Food 2010, 13, 1415–1423. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, J.H.; Lee, G.H.; Kim, H.T.; Lim, J.M.; Chae, S.W.; Chae, H.J.; Kim, H.R. p38 Mitogen-activated protein kinase is involved in endoplasmic reticulum stress-induced cell death and autophagy in human gingival fibroblasts. Biol. Pharm. Bull. 2010, 33, 545–549. [Google Scholar] [CrossRef]
- Lim, S.C.; Hahm, K.S.; Lee, S.H.; Oh, S.H. Autophagy involvement in cadmium resistance through induction of multidrug resistance-associated protein and counterbalance of endoplasmic reticulum stress WI38 lung epithelial fibroblast cells. Toxicology 2010, 276, 18–26. [Google Scholar] [CrossRef]
- Thyagarajan, A.; Jedinak, A.; Nguyen, H.; Terry, C.; Baldridge, L.A.; Jiang, J.; Sliva, D. Triterpenes from Ganoderma Lucidum induce autophagy in colon cancer through the inhibition of p38 mitogen-activated kinase (p38 MAPK). Nutr. Cancer 2010, 62, 630–640. [Google Scholar] [CrossRef]
- Colosetti, P.; Puissant, A.; Robert, G.; Luciano, F.; Jacquel, A.; Gounon, P.; Cassuto, J.P.; Auberger, P. Autophagy is an important event for megakaryocytic differentiation of the chronic myelogenous leukemia K562 cell line. Autophagy 2009, 5, 1092–1098. [Google Scholar] [CrossRef]
- Webber, J.L.; Tooze, S.A. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. Embo J. 2010, 29, 27–40. [Google Scholar] [CrossRef]
- Fritz, A.; Brayer, K.J.; McCormick, N.; Adams, D.G.; Wadzinski, B.E.; Vaillancourt, R.R. Phosphorylation of serine 526 is required for MEKK3 activity, and association with 14-3-3 blocks dephosphorylation. J. Biol. Chem. 2006, 281, 6236–6245. [Google Scholar]
- Xing, H.; Zhang, S.; Weinheimer, C.; Kovacs, A.; Muslin, A.J. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. Embo J. 2000, 19, 349–358. [Google Scholar] [CrossRef]
- Basu, A. The potential of protein kinase C as a target for anticancer treatment. Pharmacol.Ther. 1993, 59, 257–280. [Google Scholar] [CrossRef]
- Ozpolat, B.; Akar, U.; Mehta, K.; Lopez-Berestein, G. PKC delta and tissue transglutaminase are novel inhibitors of autophagy in pancreatic cancer cells. Autophagy 2007, 3, 480–483. [Google Scholar]
- Lorand, L.; Graham, R.M. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003, 4, 140–156. [Google Scholar] [CrossRef]
- Chen, J.L.; Lin, H.H.; Kim, K.J.; Lin, A.; Forman, H.J.; Ann, D.K. Novel roles for protein kinase Cdelta-dependent signaling pathways in acute hypoxic stress-induced autophagy. J. Biol. Chem. 2008, 283, 34432–34444. [Google Scholar]
- Sakaki, K.; Wu, J.; Kaufman, R.J. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J. Biol. Chem. 2008, 283, 15370–15380. [Google Scholar]
- Jiang, H.; Cheng, D.; Liu, W.; Peng, J.; Feng, J. Protein kinase C inhibits autophagy andphosphorylates LC3. Biochem. Biophys. Res. Commun. 2010, 395, 471–476. [Google Scholar] [CrossRef]
- Acs, P.; Szallasi, Z.; Kazanietz, M.G.; Blumberg, P.M. Differential activation of PKC isozymes by 14-3-3 zeta protein. Biochem. Biophys. Res. Commun. 1995, 216, 103–109. [Google Scholar] [CrossRef]
- Oriente, F.; Andreozzi, F.; Romano, C.; Perruolo, G.; Perfetti, A.; Fiory, F.; Miele, C.; Beguinot, F.; Formisano, P. Protein kinase C-alpha regulates insulin action and degradation by interacting with insulin receptor substrate-1 and 14-3-3 epsilon. J. Biol. Chem. 2005, 280, 40642–40649. [Google Scholar]
- Kim, Y.H.; Kim, Y.S.; Kang, S.S.; Noh, H.S.; Kim, H.J.; Cho, G.J.; Choi, W.S. Expression of 14-3-3 zeta and interaction with protein kinase C in the rat retina in early diabetes. Diabetologia 2005, 48, 1411–1415. [Google Scholar] [CrossRef]
- Dai, J.G.; Murakami, K. Constitutively and autonomously active protein kinase C associated with 14-3-3 zeta in the rodent brain. J. Neurochem. 2003, 84, 23–34. [Google Scholar]
- Hausser, A.; Storz, P.; Link, G.; Stoll, H.; Liu, Y.C.; Altman, A.; Pfizenmaier, K.; Johannes, F.J. Protein kinase C mu is negatively regulated by 14-3-3 signal transduction proteins. J. Biol. Chem. 1999, 274, 9258–9264. [Google Scholar]
- Meller, N.; Liu, Y.C.; Collins, T.L.; Bonnefoy-Berard, N.; Baier, G.; Isakov, N.; Altman, A. Direct interaction between protein kinase C theta (PKC theta) and 14-3-3 tau in T cells: 14-3-3 overexpression results in inhibition of PKC theta translocation and function. Mol. Cell Biol. 1996, 16, 5782–5791. [Google Scholar]
- Robinson, K.; Jones, D.; Patel, Y.; Martin, H.; Madrazo, J.; Martin, S.; Howell, S.; Elmore, M.; Finnen, M.J.; Aitken, A. Mechanism of inhibition of protein kinase C by 14-3-3 isoforms. 14- 3–3 isoforms do not have phospholipase A2 activity. Biochem. J. 1994, 29, 853–861. [Google Scholar]
- Van Der Hoeven, P.C.; Van Der Wal, J.C.; Ruurs, P.; Van Blitterswijk, W.J. Protein kinase C activation by acidic proteins including 14-3-3. Biochem. J. 2000, 347, 781–785. [Google Scholar] [CrossRef]
- Tanji, M.; Horwitz, R.; Rosenfeld, G.; Waymire, J.C. Activation of protein kinase C by purified bovine brain 14-3-3: comparison with tyrosine hydroxylase activation. J. Neurochem. 1994, 63, 1908–1916. [Google Scholar]
- Hoyer-Hansen, M.; Jaattela, M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007, 14, 1576–1582. [Google Scholar] [CrossRef]
- Davare, M.A.; Saneyoshi, T.; Guire, E.S.; Nygaard, S.C.; Soderling, T.R. Inhibition of calcium/calmodulin-dependent protein kinase kinase by protein 14-3-3. J. Biol. Chem. 2004, 279, 52191–52199. [Google Scholar]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef]
- Kamada, Y.; Yoshino, K.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef]
- Kuroyanagi, H.; Yan, J.; Seki, N.; Yamanouchi, Y.; Suzuki, Y.; Takano, T.; Muramatsu, M.; Shirasawa, T. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignmet. Genomics 1998, 51, 76–85. [Google Scholar] [CrossRef]
- Chan, E.Y.; Kir, S.; Tooze, S.A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007, 282, 25464–25474. [Google Scholar] [CrossRef]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.; Natsume, T.; Guan, J.L.; Mizushima, N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Lee, J.W.; Park, S.; Takahashi, Y.; Wang, H.G. The association of AMPK with ULK1 regulates autophagy. PLoS One 2010, e15394, 1–9. [Google Scholar]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef]
- Lawlor, M.A.; Alessi, D.R. PKB/Akt: A key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001, 114, 2903–2910. [Google Scholar]
- Wang, Y.; Yoshioka, K.; Azam, M.A.; Takuwa, N.; Sakurada, S.; Kayaba, Y.; Sugimoto, N.; Inoki, I.; Kimura, T.; Kuwaki, T.; Takuwa, Y. Class II phosphoinositide 3-kinase alpha-isoform regulates Rho, myosin phosphatase and contraction in vascular smooth muscle. Biochem. J. 2006, 394, 581–592. [Google Scholar] [CrossRef]
- Domin, J.; Harper, L.; Aubyn, D.; Wheeler, M.; Florey, O.; Haskard, D.; Yuan, M.; Zicha, D. The class II phosphoinositide 3-kinase PI3K-C2beta regulates cell migration by a PtdIns3P dependent mechanism. J. Cell Physiol. 2005, 205, 452–462. [Google Scholar] [CrossRef]
- Backer, J.M. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 2008, 410, 1–17. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Waddell, S.; Gaugel, A.; Frickey, T.; Lupas, A.; Nordheim, A. WIPI- 1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004, 23, 9314–9325. [Google Scholar] [CrossRef]
- Polson, H.E.; de Lartigue, J.; Rigden, D.J.; Reedijk, M.; Urbe, S.; Clague, M.J.; Tooze, S.A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010, 6. [Google Scholar]
- Krick, R.; Tolstrup, J.; Appelles, A.; Henke, S.; Thumm, M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006, 580, 4632–4638. [Google Scholar] [CrossRef]
- Ghosh, P.; Wu, M.; Zhang, H.; Sun, H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle 2008, 7, 373–381. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1- phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Pozuelo-Rubio, M. Regulation of autophagic activity by 14-3-3zeta proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ. 2011, 18, 479–492. [Google Scholar] [CrossRef]
- Wang, B.; Lin, S.; Lin, W.-C. 14-3-3tau regulates Beclin 1 and is required for autophagy. 2010, 5, e10409, 1–10. [Google Scholar]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Lee, J.S.; Li, Q.; Lee, J.Y.; Lee, S.H.; Jeong, J.H.; Lee, H.R.; Chang, H.; Zhou, F.C.; Gao, S.J.; Liang, C.; Jung, J.U. FLIP-mediated autophagy regulation in cell death control. Nat. Cell. Biol. 2009, 11, 1355–1362. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pozuelo-Rubio, M. 14-3-3 Proteins are Regulators of Autophagy. Cells 2012, 1, 754-773. https://doi.org/10.3390/cells1040754
Pozuelo-Rubio M. 14-3-3 Proteins are Regulators of Autophagy. Cells. 2012; 1(4):754-773. https://doi.org/10.3390/cells1040754
Chicago/Turabian StylePozuelo-Rubio, Mercedes. 2012. "14-3-3 Proteins are Regulators of Autophagy" Cells 1, no. 4: 754-773. https://doi.org/10.3390/cells1040754



