Ciliary/Flagellar Protein Ubiquitination
Abstract
:1. Introduction

2. Ubiquitin Conjugating System is Associated with Cilia
3. Flagellar Protein Ubiquitination and Flagellar Disassembly
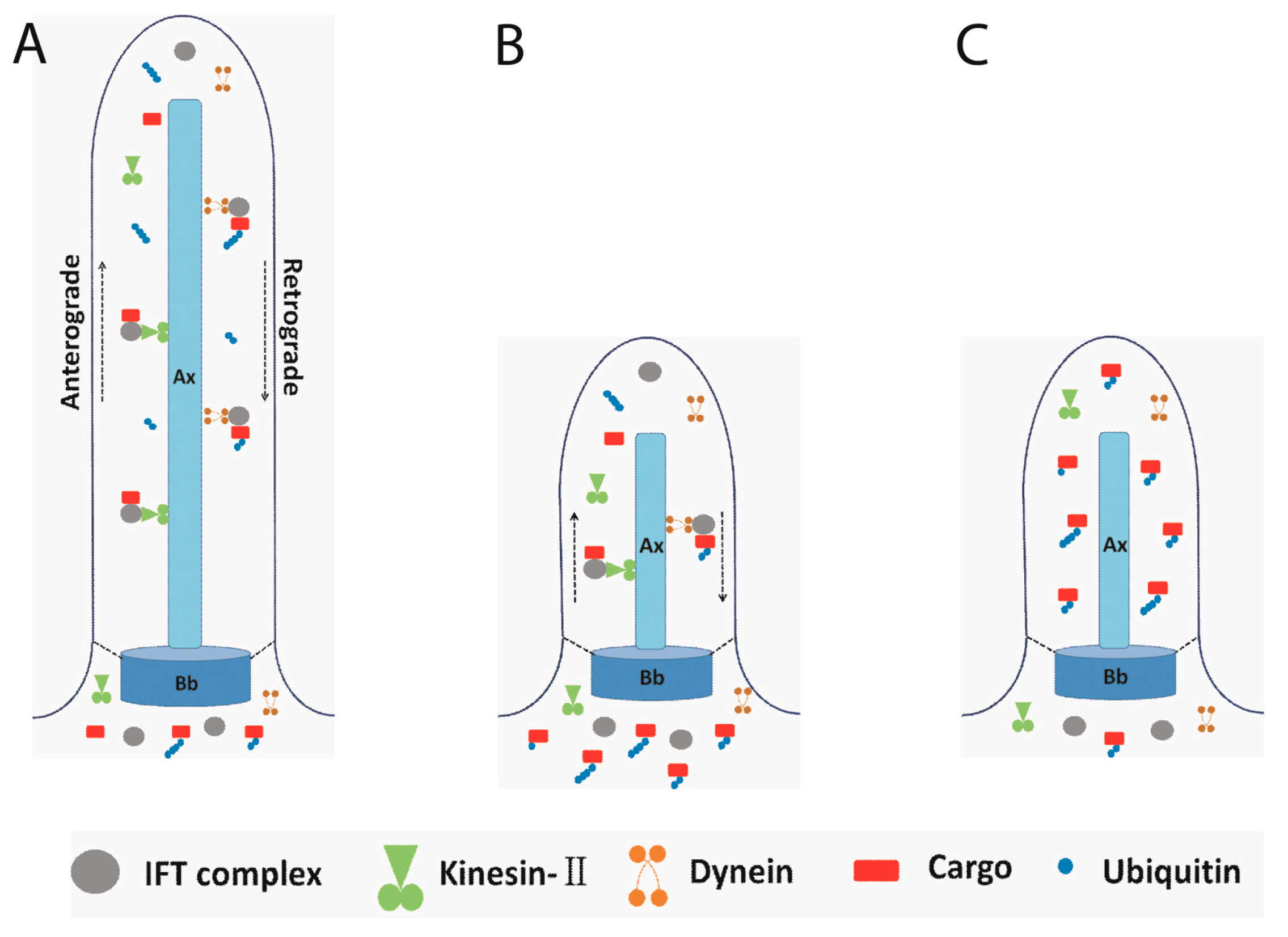
4. Flagellar Protein Ubiquitination and Signal Transduction
5. Flagellar Protein Ubiquitination and Spermatogenesis
6. Ciliary Protein Ubiquitination in Primary Cilia
7. Summary and Future Research Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.S.; Ben-Shahar, Y.; Moninger, T.O.; Kline, J.N.; Welsh, M.J. Motile cilia of human airway epithelia are chemosensory. Science 2009, 325, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Pazour, G.J.; Witman, G.B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003, 15, 105–110. [Google Scholar] [CrossRef]
- Singla, V.; Reiter, J.F. The primary cilium as the cell's antenna: Signaling at a sensory organelle. Science 2006, 313, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.L.; Witman, G.B. Intraflagellar transport. Nature reviews. Nat. Rev. Mol. Cell Biol. 2002, 3, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Tilley, A.E.; Walters, M.S.; Shaykhiev, R.; Crystal, R.G. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 2015, 77, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.K. Polycystic kidney disease: Pathogenic missense mutations result in defective trafficking of polycystin-1 to cilia. Nat. Rev. Nephrol. 2015, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Klena, N.T.; Gabriel, G.C.; Liu, X.; Kim, A.J.; Lemke, K.; Chen, Y.; Chatterjee, B.; Devine, W.; Damerla, R.R.; et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 2015, 521, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Dell, K.M. The role of cilia in the pathogenesis of cystic kidney disease. Curr. Opin. Pediatr. 2015, 27, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, F.; Benzing, T.; Katsanis, N. Ciliopathies. N. Engl. J. Med. 2011, 364, 1533–1543. [Google Scholar] [PubMed]
- Dentler, W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol. 2005, 170, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Yanagisawa, H.A.; Liu, Z.; Shibuya, R.; Hirono, M.; Kamiya, R. A conserved flagella-associated protein in Chlamydomonas, FAP234, is essential for axonemal localization of tubulin polyglutamylase TTLL9. Mol. Biol. Cell 2014, 25, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kozminski, K.G.; Diener, D.R.; Rosenbaum, J.L. High level expression of nonacetylatable alpha-tubulin in Chlamydomonas reinhardtii. Cell Motil. Cytoskel. 1993, 25, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Dossou, S.J.; Bre, M.H.; Hallworth, R. Mammalian cilia function is independent of the polymeric state of tubulin glycylation. Cell Motil. Cytoskel. 2007, 64, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Diener, D.R.; Rosenbaum, J.L. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 2009, 186, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Li, G.; Pan, J. Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol. 2009, 94, 333–346. [Google Scholar] [PubMed]
- Wilson, N.F.; Lefebvre, P.A. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 2004, 3, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Meng, D.; Wang, L.; Bei, S.; Snell, W.J.; Pan, J. Activation loop phosphorylation of a protein kinase is a molecular marker of organelle size that dynamically reports flagellar length. Proc. Natl. Acad. Sci. USA 2013, 110, 12337–12342. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liang, Y.; He, W.; Pan, J. Cilia disassembly with two distinct phases of regulation. Cell Rep. 2015, 10, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Naumann-Busch, B.; Wang, L.; Specht, M.; Scholz, M.; Trompelt, K.; Hippler, M. Protein phosphorylation is a key event of flagellar disassembly revealed by analysis of flagellar phosphoproteins during flagellar shortening in Chlamydomonas. J. Proteome Res. 2011, 10, 3830–3839. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.J.; Ulland, M.; Sloboda, R.D. A protein methylation pathway in Chlamydomonas flagella is active during flagellar resorption. Mol. Biol. Cell 2008, 19, 4319–4327. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Sato, S.; Nakamura, K.; Ostrowski, L.E.; Setou, M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc. Natl. Acad. Sci. USA 2010, 107, 10490–10495. [Google Scholar] [CrossRef] [PubMed]
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 2009, 458, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Corbit, K.C.; Aanstad, P.; Singla, V.; Norman, A.R.; Stainier, D.Y.; Reiter, J.F. Vertebrate Smoothened functions at the primary cilium. Nature 2005, 437, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- Welchman, R.L.; Gordon, C.; Mayer, R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005, 6, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [PubMed]
- Pazour, G.J.; Agrin, N.; Leszyk, J.; Witman, G.B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005, 170, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, O.; Malinowska, A.; Klotz, C.; Sperling, L.; Dadlez, M.; Koll, F.; Cohen, J. Cildb: A knowledgebase for centrosomes and cilia. Database (Oxford) 2009, 2009, bap022. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Huang, K. Analysis of flagellar protein ubiquitination. Methods Enzymol. 2013, 524, 59–73. [Google Scholar] [PubMed]
- Liu, Q.; Tan, G.; Levenkova, N.; Li, T.; Pugh, E.N., Jr.; Rux, J.J.; Speicher, D.W.; Pierce, E.A. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 2007, 6, 1299–1317. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Thompson, J.; Yates, J.R., 3rd; Marshall, W.F. Proteomic analysis of mammalian primary cilia. Curr. Biol. 2012, 22, 414–419. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.; O’Bryan, M.K. Microtubules and spermatogenesis. Semin. Cell Dev. Biol. 2014, 30, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hermo, L.; Pelletier, R.M.; Cyr, D.G.; Smith, C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 3: Developmental changes in spermatid flagellum and cytoplasmic droplet and interaction of sperm with the zona pellucida and egg plasma membrane. Microsc. Res. Tech. 2010, 73, 320–363. [Google Scholar] [CrossRef] [PubMed]
- Kierszenbaum, A.L. Sperm axoneme: A tale of tubulin posttranslation diversity. Mol. Reprod. Dev. 2002, 62, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Manku, G.; Culty, M.; Wing, S.S. Ubiquitin-proteasome system in spermatogenesis. Adv. Exp. Med. Biol. 2014, 759, 181–213. [Google Scholar] [PubMed]
- Mochida, K.; Tres, L.L.; Kierszenbaum, A.L. Structural features of the 26S proteasome complex isolated from rat testis and sperm tail. Mol. Reprod. Dev. 2000, 57, 176–184. [Google Scholar] [CrossRef]
- Markelewicz, R.J., Jr.; Hall, S.J.; Boekelheide, K. 2,5-Hexanedione and carbendazim coexposure synergistically disrupts rat spermatogenesis despite opposing molecular effects on microtubules. Toxicol. Sci. 2004, 80, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ito, K.; Iyengar, P.V.; Hirose, S.; Nakamura, N. MARCH7 E3 ubiquitin ligase is highly expressed in developing spermatids of rats and its possible involvement in head and tail formation. Histochem. Cell Biol. 2013, 139, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, P.V.; Hirota, T.; Hirose, S.; Nakamura, N. Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J. Biol. Chem. 2011, 286, 39082–39090. [Google Scholar] [CrossRef] [PubMed]
- Yasuzumi, G.; Matano, Y.; Asai, T.; Nagasaka, M.; Yasuzumi, F. Spermatogenesis in animals as revealed by electron microscopy. XXII. Development of nuclei and cytoplasmic microtubules in the grasshopper spermatids. Z. Zellforsch. Mikrosk. Anat. 1971, 115, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Yasuzumi, G.; Sugioka, T. Spermatogenesis in animals as revealed by electron microscopy. XXI. Microkaryosomes and microtubules appearing during spermiogenesis of the lovebird Uroloncha striata var. domestica flower. Z. Zellforsch. Mikrosk. Anat. 1971, 114, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Fouquet, J.; Kann, M.; Soues, S.; Melki, R. ARP1 in Golgi organisation and attachment of manchette microtubules to the nucleus during mammalian spermatogenesis. J. Cell Sci. 2000, 113, 877–886. [Google Scholar] [PubMed]
- Lerer-Goldshtein, T.; Bel, S.; Shpungin, S.; Pery, E.; Motro, B.; Goldstein, R.S.; Bar-Sheshet, S.I.; Breitbart, H.; Nir, U. TMF/ARA160: A key regulator of sperm development. Dev. Biol. 2010, 348, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Ugwunna, S.C.; Foor, W.E. The function of microtubules during spermatogenesis of Ancylostoma caninum. J. Parasitol. 1982, 68, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Kawakami, Y.; Kiyono, T.; Yonemura, S.; Kawamura, Y.; Era, S.; Matsuzaki, F.; Goshima, N.; Inagaki, M. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat. Commun. 2014, 5, 5081. [Google Scholar] [CrossRef] [PubMed]
- Villumsen, B.H.; Danielsen, J.R.; Povlsen, L.; Sylvestersen, K.B.; Merdes, A.; Beli, P.; Yang, Y.G.; Choudhary, C.; Nielsen, M.L.; Mailand, N.; et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013, 32, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Pabla, N.; Huang, S.; Dong, Z. Nek1 phosphorylates Von Hippel-Lindau tumor suppressor to promote its proteasomal degradation and ciliary destabilization. Cell Cycle 2013, 12, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Eguether, T.; Ermolaeva, M.A.; Zhao, Y.; Bonnet, M.C.; Jain, A.; Pasparakis, M.; Courtois, G.; Tassin, A.M. The deubiquitinating enzyme CYLD controls apical docking of basal bodies in ciliated epithelial cells. Nat. Commun. 2014, 5, 4585. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, H.; Wang, Q.; Huang, K. Ciliary/Flagellar Protein Ubiquitination. Cells 2015, 4, 474-482. https://doi.org/10.3390/cells4030474
Long H, Wang Q, Huang K. Ciliary/Flagellar Protein Ubiquitination. Cells. 2015; 4(3):474-482. https://doi.org/10.3390/cells4030474
Chicago/Turabian StyleLong, Huan, Qiyu Wang, and Kaiyao Huang. 2015. "Ciliary/Flagellar Protein Ubiquitination" Cells 4, no. 3: 474-482. https://doi.org/10.3390/cells4030474




