MicroRNAs and Molecular Mechanisms of Neurodegeneration
Abstract
:Abbreviations
| Ago2 = Argonaute-2 | ALS = Amyotrophic Lateral Sclerosis |
| APP = Amyloid precursor protein | BACE1 = beta-site APP-cleavage enzyme 1 |
| BTBD3 = BTB (POZ) domain containing 3 | COL2A1 = Collagen, type II, alpha 1 |
| CoREST = REST Corepressor | COXIV = cytochrome c oxidase IV subunit |
| dAgo1 = Drosophila Argonaute-1 | DAT = Dopamine transporter |
| DJ1 = Parkin-7 | E2F1/DP = E2F transcription factor 1 |
| EAAT2 = excitatory amino acid transporter 2 | TDP-43 = TAR DNA-binding protein 43 |
| GALC = galactosylceramide | GLT-1 = glutamate transporter |
| hAgo2 = human Argonaute-2 | HDAC4 = Histone Deacetylase 4 |
| HTT = Huntingtin | LSD = Lysosomal Disease |
| NEFL = Neurofilament light polypeptide | NfKB = nuclear factor kappa BNPC = Niemann Pick cells |
| PACT = protein activator of PKR | TBP = Tata Binding Protein |
| phospho-4E-BP1 = 4E binding protein1 | Pitx3 = paired-like homeodomain transcription factor 3 |
| PTBP2 = Polypyrimidine tract binding protein 2 | RLC = regulatory light chain |
| SH-SY5Y = Human Neuroblastoma Cells Line | SIRT1 = sirtuin 1 |
| SNP = single-nucleotide polymorphism | SPT = Serine palmitoyltransferase |
| TAp73 = Tumor protein p73 | ERK1 = Extracellular signal-regulated kinase 1FUS/TLS = fused in sarcoma/traslocated in liposarcoma |
| PGC-1α = Peroxisome Proliferator—Activated Receptor Gamma Coactivator 1 | TGFBI = transforming growth factor, beta 1TLR-7 = Toll-like receptor 7 |
| TRBP = Tar RNA binding proteinTRIM2 = tripartite motif containing 2 |
1. Biogenesis and Role of microRNAs
1.1. Canonical Function of microRNAs
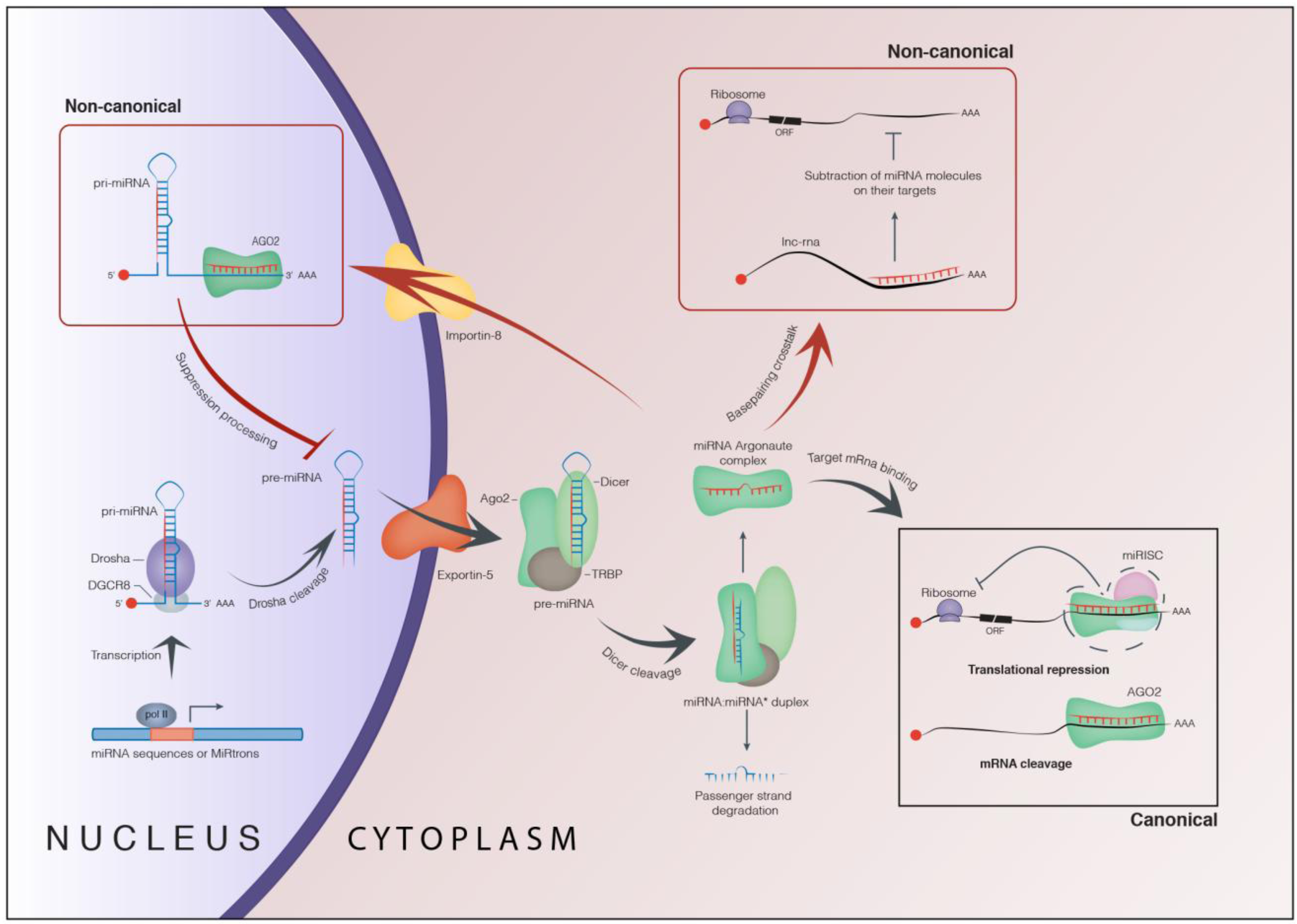
1.2. Non-Canonical Function of microRNAs
2. MicroRNAs and Neurodegeneration
| microRNA | Neurodegenerative Disease | Molecular Target | Effects | Reference |
|---|---|---|---|---|
| miR-15 | AD | ERK1 and Tau | ERK1 and Tau regulation | [54] |
| miR-16 | AD | APP | Overexpression reduced APP level | [55] |
| ERK1 and Tau | ERK1 and Tau regulation | [54] | ||
| miR-106a | AD | APP | APP repression | [56] |
| miR-106b | AD | APP | Aberrantly expressed in APPswe/PSE9 mice | [57] |
| miR-107 | AD | BACE1 | Downregulated. | [58] |
| Repression of Cofilin translation, a component of rod-like actin structures in the AD brain. | ||||
| miR-124 | AD | BACE1 | Suppressed induces over expression of BACE1 | [59] |
| miR-132 | AD | PTBP2 | Neuronal splicing regulator of Tau Exon 10 | [54] |
| miR-137 | AD | SPT | SPT and in turn Aβ levels up-regulate | [60] |
| miR-153 | AD | APP | Downregulated in modest AD pathology | [56] |
| miR-195 | AD | BACE1 | Overexpressed decreased BACE1 protein level | [61] |
| miR-497 | AD | ERK1 and Tau | ERK1 and Tau regulation | [54] |
| miR-520c | AD | APP | APP repression | [56] |
| Let-7b | AD | TLR-7 | Induce Toll-like receptor 7 activation | [62] |
| miR-7 | PD | α-synuclein mRNA | It can represses α-synuclein protein levels collaborating with miR-153 | [63] |
| miR-133b | PD | Pitx3 | Downregulated in PD brain tissue | [64] |
| miR-34b/c | PD | SH-SY5Y dopaminergic neuron | Downregulated | [65] |
| miR-let7 | PD | LRRK2 | Regulation of Drosophila e2f1 protein synthesis: repressed expression | [66] |
| miR-184 | PD | LRRK2 | Regulation of dp messenger RNAs synthesis: repressed expression | [66] |
| miR-433 | PD | SNP rs12720208 in the 3' UTR | Increased FGF20 expression and upregulation of alpha-synuclein | [67] |
| miR-9/miR-9* | HD | REST/COREST | Downregulated. Double negative feedback loop between the REST silencing complex and the miRNAs it regulates | [68] |
| [69] | ||||
| miR-29c | HD | REST | Downregulated | [70] |
| miR-34b | HD | p53 | Mysregulated causing by mHTT accumulation. Overexpressed in plasma of HD patients | [71] |
| miR-124 | HD | REST | Downregulated leads to an increases of their target level | [68] |
| miR-125b | HD | HTT | Downregulated | [72] |
| miR-222 | HD | REST | Downregulated. | [70] |
| miR-132 | HD | REST | Downregulated. Neurite sprouting | [68] |
| miR-135 | HD | REST | Downregulated | [68] |
| miR-137 | HD | REST | Aberrantly repressed directly mediated by REST | [73] |
| miR-146a | HD | TBP | Regulation of TBP by miR-146a may contribute to HD pathogenesis. Generally downregulated | [72] |
| miR-150 | HD | HTT | Downregulated | [72] |
| miR-153 | HD | REST | Downregulated | [73] |
| miR-200a | HD | Genes regulating synaptic function, neurodevelopment, and neuronal survival | Upregulated. Perturbed expression in HD patients. | [74] |
| miR-200c | [74] | |||
| miR-9 | ALS | NEFL | Downregulated | [75] |
| miR-23a | ALS | PGC-1 | Upregulated. It can reduce PGC-1α signalling, cytochome-b and COXIV protein levels | [76] |
| miR-29b | ALS | p53 | Upregulated | [76] |
| miR-124 | ALS | EAAT2/GLT1 | Indirect miR-124a-mediated regulation of GLT1 expression from neurons to astrocytes | [77] |
| miR-206 | ALS | HDAC4 | Upregulated in ALS end stage model to regenerate damaged neuromuscular synapses by HDAC4 reinnervation via | [78] |
| miR-455 | ALS | COL2A1 | Upregulated in skeletal muscles of ALS patients | [76] |
| Let-7b | ALS | TDP-43 | Downregulated | [79] |
| miR-663 | ALS | TDP-43 | Upregulated | [79] |
| miR-126 | LSD | GALC | Expressed in HSCs but not in differentiated cells | [80] |
| miR-130 | [80] | |||
| miR-196a | NPC | Lipid biosynthesis associated genes | Upregulated | [81] |
| miR-196b | NPC | Lipid biosynthesis associated genes | Upregulated | [81] |
| miR-296 | NPC | Lipid biosynthesis associated genes | Upregulated | [81] |
| miR-98 | NPC | Lipid biosynthesis associated genes | Downregulated.Lipid biosynthesis associated | [81] |
| miR-143 | NPC | Lipid biosynthesis associated genes | Downregulated. | [81] |
| Lipid biosynthesis associated |
2.1. MicroRNAs and Alzheimer’s Disease

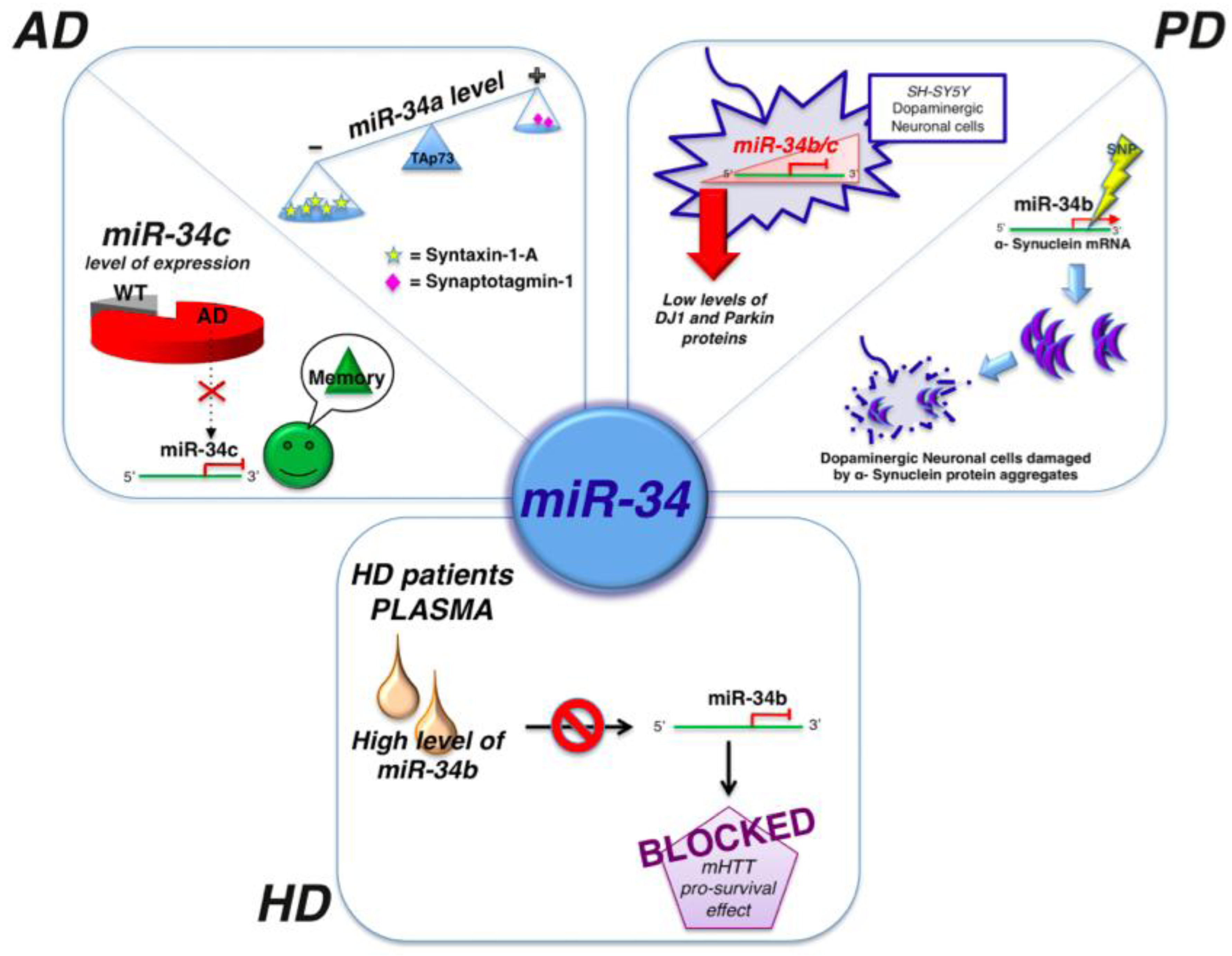
2.2. MicroRNAs and Parkinson’s Disease
2.3. MicroRNAs and Hungtinton’s Disease
2.4. MicroRNAs and Amyotrophic Lateral Sclerosis
2.5. MicroRNAs and Lysosomal Storage Disorders
3. Concluding Remarks
Acknowledgements
References
- Bartel, D.P. microRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. Posttranscriptionalregulation of microRNAbiogenesis in animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanism of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; Kim, V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-trascriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursor that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells ewpress endogenous shRNAs, siRNAs, and other Microprocesso-indipendent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Saraiya, A.A.; Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef]
- Czech, B.; Zhou, R.; Erlich, Y.; Brennecke, J.; Binari, R.; Villalta, C.; Gordon, A.; Perrimon, N.; Hannon, G.J. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell 2009, 36, 445–456. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Hartig, J.V.; Siomi, H.; Siomi, M.C. Molecular mechanism that funnel RNA precursor into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA 2010, 16, 506–515. [Google Scholar] [CrossRef]
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Lainé, S.; Sack, R.; Gatignol, A.; Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005, 6, 961–967. [Google Scholar] [CrossRef]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404. [Google Scholar] [CrossRef]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Zeng, Y.; Wagner, E.J.; Cullen, B.R. Both natural designed micro RNAs can inhibit expression of cognate mRNAs when expressed in human cells. Mol. Cell 2002, 9, 1327–1333. [Google Scholar] [CrossRef]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef]
- Djuranovic, S.; Nahvi, A.; Green, R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012, 336, 237–240. [Google Scholar] [CrossRef]
- Shukla, G.C.; Singh, J.; Barik, S. microRNAs: Processing, maturation, target recognition and regulatory functions. Mol. Cell Pharmacol. 2011, 3, 83–92. [Google Scholar]
- Kim, D.H.; Saetrom, P.; Snøve, O., Jr.; Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar]
- Chen, X.; Liang, H.; Zhang, C.Y.; Zen, K. miRNA regulates noncoding RNA: A noncanonical function model. Trends Biochem. Sci. 2012, 37, 457–459. [Google Scholar] [CrossRef]
- Xia, J.; Joyce, C.E.; Bowcock, A.M.; Zhang, W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum. Mol. Genet. 2013, 22, 737–748. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–368. [Google Scholar]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al.; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef]
- Karreth, F.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.; Rust, A.G.; DeNicola, G.; Webster, K.A.; Weiss, D.; Perez-Mancera, P.A.; et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011, 147, 382–395. [Google Scholar] [CrossRef]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef]
- Orlacchio, A.; Bernardi, G.; Orlacchio, A.; Martino, S. Stem cells: An overview of the current status of therapies for central and peripheral nervous system diseases. Curr. Med. Chem. 2010, 17, 595–608. [Google Scholar] [CrossRef]
- Orlacchio, A.; Bernardi, G.; Orlacchio, A.; Martino, S. Stem cells and neurological diseases. Discov. Med. 2010, 9, 546–553. [Google Scholar]
- Abe, M.; Bonini, N.M. microRNAs and neurodegeneration: Role and impact. Trends Cell. Biol. 2013, 23, 30–36. [Google Scholar] [CrossRef]
- Costa, V.; Esposito, R.; Aprile, M.; Ciccodicola, A. Non-coding RNA and pseudogenes in neurodegenerative diseases: “The (un)Usual Suspects”. Front. Genet. 2012, 3, 231. [Google Scholar]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef]
- Martino, S.; di Girolamo, I.; Orlacchio, A.; Datti, A.; Orlacchio, A. microRNA implications across neurodevelopment and neuropathology. J. Biomed. Biotechnol. 2009, 2009, 654346. [Google Scholar]
- Arevalo-Rodriguez, I.; Pedraza, O.L.; Rodríguez, A.; Sánchez, E.; Gich, I.; Solà, I.; Bonfill, X.; Alonso-Coello, P. Alzheimer’s disease dementia guidelines for diagnostic testing: A systematic review. Am. J. Alzheimers Dis. Other Demen. 2013, 28, 111–119. [Google Scholar]
- Surmeier, D.J.; Sulzer, D. The pathology roadmap inParkinson disease. Prion 2013, 7, 85–91. [Google Scholar] [CrossRef]
- Al-Chalabi, A.; Jones, A.; Troakes, C.; King, A.; Al-Sarraj, S.; van den Berg, L.H. The genetics and neuropathology ofamyotrophic lateral sclerosis. Acta Neuropathol. 2012, 124, 339–352. [Google Scholar] [CrossRef]
- Tierney, T.S.; Vasudeva, V.S.; Weir, S.; Hayes, M.T. Neuromodulation for neurodegenerative conditions. Front. Biosci. (Elite Ed.) 2013, 5, 490–499. [Google Scholar]
- Platt, F.M.; Boland, B.; van der Spoel, A.C. The cell biology of disease: Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J. Cell. Biol. 2012, 199, 723–734. [Google Scholar] [CrossRef]
- Hébert, S.S.; Sergeant, N.; Buée, L. microRNAs and the regulation oftau metabolism. Int. J. Alzheimers Dis. 2012, 2012, 406561. [Google Scholar]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar] [CrossRef]
- Long, J.M.; Lahiri, D.K. Current drug targets for modulating Alzheimer’s amyloid precursor protein: Role of specific micro-RNA species. Curr. Med. Chem. 2011, 18, 3314–3321. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Zong, Y.; Xu, Y.; Deng, W.; Zhu, H.; Liu, Y.; Ma, C.; Huang, L.; Zhang, L.; Qin, C. miR-106b is aberrantly expressed in a double transgenic mouse model for Alzheimer’s desease targets TGF-B typeII receptor. Brain Res. 2010, 1357, 166–174. [Google Scholar]
- Yao, J.; Hennessey, T.; Flynt, A.; Lai, E.; Beal, M.F.; Lin, M.T. microRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One 2010, 5, e15546. [Google Scholar]
- Fang, M.; Wang, J.; Zhang, X.; Geng, Y.; Hu, Z.; Rudd, J.A.; Ling, S.; Chen, W.; Han, S. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol. Lett. 2011, 209, 94–105. [Google Scholar]
- Geekiyanage, H.; Chan, C. microRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J. Neurosci. 2011, 31, 14820–14830. [Google Scholar] [CrossRef]
- Zhu, H.C.; Wang, L.M.; Wang, M.; Song, B.; Tan, S.; Teng, J.F.; Duan, D.X. microRNA-195 downregulates Alzheimer’sdisease amyloid-beta production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef]
- Lehmann, S.M. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar]
- Junn, E.; Lee, K.W.; Jeong, B.S.; Chan, T.W.; Im, J.Y.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl. Acad. Sci. USA 2009, 106, 13052–13057. [Google Scholar]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007, 317, 1220–1224. [Google Scholar]
- Miñones-Moyano, E.; Porta, S.; Escaramís, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Martí, E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. [Google Scholar]
- Gehrke, S.; Imai, Y.; Sokol, N.; Lu, B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 2010, 466, 637–641. [Google Scholar] [CrossRef]
- De Mena, L.; Coto, E.; Cardo, L.F.; Díaz, M.; Blázquez, M.; Ribacoba, R.; Salvador, C.; Pastor, P.; Samaranch, L.; Moris, G.; et al. Analysis of the Micro-RNA-133 and PITX3 genes in Parkinson’s disease. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153B, 1234–1239. [Google Scholar]
- Johnson, R.; Zuccato, C.; Belyaev, N.D.; Guest, D.J.; Cattaneo, E.; Buckley, N.J. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 2008, 29, 438–445. [Google Scholar]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008, 28, 14341–14346. [Google Scholar]
- Lee, S.T.; Chu, K.; Im, W.S.; Yoon, H.J.; Im, J.Y.; Park, J.E.; Park, K.H.; Jung, K.H.; Lee, S.K.; Kim, M.; et al. Altered microRNA regulation in Huntington’s disease models. Exp. Neurol. 2011, 227, 172–179. [Google Scholar]
- Gaughwin, P.M.; Ciesla, M.; Lahiri, N.; Tabrizi, S.J.; Brundin, P.; Björkqvist, M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum. Mol. Genet. 2011, 20, 2225–2237. [Google Scholar] [CrossRef]
- Ghose, J.; Sinha, M.; Das, E.; Jana, N.R.; Bhattacharyya, N.P. Regulation of miR-146a by RelA/NFkB and p53 in STHdh(Q111)/Hdh(Q111) cells, a cell model of Huntington’s disease. PLoS One 2011, 6, e23837. [Google Scholar]
- Soldati, C.; Bithell, A.; Johnston, C.; Wong, K.Y.; Stanton, L.W.; Buckley, N.J. Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington’s disease. J. Neurochem. 2012, 124, 418–430. [Google Scholar]
- Jin, J.; Cheng, Y.; Zhang, Y.; Wood, W.; Peng, Q.; Hutchison, E.; Mattson, M.P.; Becker, K.G.; Duan, W. Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J. Neurochem. 2012, 123, 477–490. [Google Scholar] [CrossRef]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.W.; Wills, A.M.; Wirguin, I.; et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar]
- Russell, A.P.; Wada, S.; Vergani, L.; Hock, M.B.; Lamon, S.; Léger, B.; Ushida, T.; Cartoni, R.; Wadley, G.D.; Hespel, P.; et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 2012, 49C, 107–117. [Google Scholar]
- Morel, L.; Regan, M.; Higashimori, H.; Ng, S.K.; Esau, C.; Vidensky, S.; Rothstein, J.; Yang, Y. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J. Biol. Chem. 2013, 288, 7105–7116. [Google Scholar] [CrossRef]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. microRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar]
- Buratti, E.; de Conti, L.; Stuani, C.; Romano, M.; Baralle, M.; Baralle, F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010, 277, 2268–2281. [Google Scholar] [CrossRef]
- Gentner, B.; Visigalli, I.; Hiramatsu, H.; Lechman, E.; Ungari, S.; Giustacchini, A.; Schira, G.; Amendola, M.; Quattrini, A.; Martino, S.; et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2010, 2, 58ra84. [Google Scholar] [CrossRef]
- Ozsait, B.; Komurcu-Bayrak, E.; Levula, M.; Erginel-Unaltuna, N.; Kähönen, M.; Rai, M.; Lehtimäki, T.; Laaksonen, R. Niemann-Pick type C fibroblasts have a distinct microRNA profile related to lipid metabolism and certain cellular components. Biochem. Biophys. Res. Commun. 2010, 403, 316–321. [Google Scholar] [CrossRef]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar]
- Selkoe, D.; Mandelkow, E.; Holtzman, D. Deciphering Alzheimer disease. Cold Spring Harb. Perspect Med. 2012, 2, a011460. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef]
- Long, J.M.; Ray, B.; Lahiri, D.K. microRNA-153 physiolofically inhibits expression of amyloid-B precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 2012, 287, 31298–31310. [Google Scholar]
- Liang, C.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Deng, W.; Liu, Y.; Qin, C. microRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012, 1455, 103–113. [Google Scholar] [CrossRef]
- Zovoilis, A.; Agbemenyah, H.Y.; Agis-Balboa, R.C.; Stilling, R.M.; Edbauer, D.; Rao, P.; Farinelli, L.; Delalle, I.; Schmitt, A.; Falkai, P.; et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011, 30, 4299–4308. [Google Scholar] [CrossRef]
- Wang, X.; Liu, P.; Zhu, H.; Xu, Y.; Ma, C.; Dai, X.; Huang, L.; Liu, Y.; Zhang, L.; Qin, C. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res. Bull. 2009, 80, 268–273. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Killick, R.; Candi, E.; Sayan, B.S.; Rivetti di Val Cervo, P.; Nicotera, P.; McKeon, F.; Knight, R.A.; Mak, T.W.; et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. USA 2011, 108, 21093–21098. [Google Scholar]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; de Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Hirano, A. Hirano bodies and related neuronal inclusions. Neuropathol. Appl. Neurobiol. 1994, 20, 3–11. [Google Scholar] [CrossRef]
- Schonrock, N.; Humphreys, D.T.; Preiss, T.; Götz, J. Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-β. J. Mol. Neurosci. 2012, 46, 324–335. [Google Scholar]
- Hoehn, M.; Yahr, M.D. Parkinsonism: Onset, progression, and mortality. Neurology 2001, 57, 11–26. [Google Scholar]
- Reinhardt, A.; Feuillette, S.; Cassar, M.; Callens, C.; Thomassin, H.; Birman, S.; Lecourtois, M.; Antoniewski, C.; Tricoire, H. Lack of miRNA misregulation at early pathological stages in drosophila neurodegenerative disease models. Front. Genet. 2012, 3, 226. [Google Scholar]
- Shtilbans, A.; Henchcliffe, C. Biomarkers in Parkinson’s disease: An update. Curr. Opin. Neurol. 2012, 25, 460–465. [Google Scholar] [CrossRef]
- Kapushesky, M.; Adamusiak, T.; Burdett, T.; Culhane, A.; Farne, A.; Filippov, A.; Holloway, E.; Klebanov, A.; Kryvych, N.; Kurbatova, N.; et al. Gene Expression Atlas update—A value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2012, 40, D1077–D1081. [Google Scholar] [CrossRef]
- Thomas, B.; Beal, M.F. Parkinson’s disease. Hum. Mol. Genet. 2007, 16, R183–R194. [Google Scholar] [CrossRef]
- Khodr, C.E.; Pedapati, J.; Han, Y.; Bohn, M.C. Inclusion of a portion of the native SNCA 3'UTR reduces toxicity of human S129A SNCA on striatal-projecting dopamine neurons in rat substantia nigra. Dev. Neurobiol. 2012, 72, 906–917. [Google Scholar]
- Doxakis, E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 2010, 285, 12726–12734. [Google Scholar] [CrossRef]
- Santosh, P.S.; Arora, N.; Sarma, P.; Pal-Bhadra, M.; Bhadra, U. Interaction map and selection of microRNA targets in Parkinson’s disease-related genes. J. Biomed. Biotechnol. 2009, 2009, 363145. [Google Scholar]
- Wang, G.; van der Walt, J.M.; Mayhew, G.; Li, Y.J.; Züchner, S.; Scott, W.K.; Martin, E.R.; Vance, J.M. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008, 82, 283–289. [Google Scholar] [CrossRef]
- Hébert, S.S.; de Strooper, B. Molecular biology miRNAs in neurodegeneration. Science 2007, 317, 1179–1180. [Google Scholar] [CrossRef]
- Heyer, M.P.; Pani, A.K.; Smeyne, R.J.; Kenny, P.J.; Fleng, G. Normal midbrain dopaminergic neuron development and function in miR133b mutant mice. J. Neurosci. 2012, 32, 10887–10894. [Google Scholar]
- Margis, R.; Margis, R.; Rieder, C.R. Identification of blood microRNAs associated to Parkinson’s disease. J. Biotechnol. 2011, 152, 96–101. [Google Scholar]
- Mouradian, M.M. MicroRNAs in Parkinson’s disease. Neurobiol. Dis. 2012, 46, 279–284. [Google Scholar] [CrossRef]
- Gascon, E.; Gao, F.B. Cause or effect: Misregulation of microRNA pathways in Neurodegeneration. Front. Neurosci. 2012, 6, 48. [Google Scholar]
- Ha, T.Y. microRNAs in human diseases: From autoimmune diseases to skin. Psychiatr. Neurodegener. Dis. Immune Netw. 2011, 11, 227–244. [Google Scholar]
- Fiszer, A.; Olejniczak, M.; Switonski, P.M.; Wroblewska, J.P.; Wisniewska-Kruk, J.; Mykowska, A.; Krzyzosiak, W.J. An evaluation of oligonucleotide-based therapeutic strategies for polyQ diseases. BMC Mol. Biol. 2012, 13, 6. [Google Scholar]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar]
- Hu, J.; Liu, J.; Corey, D.R. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010, 17, 1183–1188. [Google Scholar]
- Hodges, A.; Strand, A.D.; Aragaki, A.K.; Kuhn, A.; Sengstag, T.; Hughes, G.; Elliston, L.A.; Hartog, C.; Goldstein, D.R.; Thu, D.; et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006, 15, 965–977. [Google Scholar]
- Buckley, N.J.; Johnson, R.; Zuccato, C.; Bithell, A.; Cattaneo, E. The role of REST in transcriptional and epigenetic dysregulation in Huntington’s disease. Neurobiol. Dis. 2010, 39, 28–39. [Google Scholar]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. microRNA-34b and MicroRNA-34c are targets of P53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef]
- Morlando, M.; Dini Modigliani, S.; Torrelli, G.; Rosa, A.; di Carlo, V.; Caffarelli, E.; Bozzoni, I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012, 31, 4502–4510. [Google Scholar] [CrossRef]
- Butovsky, O.; Siddiqui, S.; Gabriely, G.; Lanser, A.J.; Dake, B.; Murugaiyan, G.; Doykan, C.E.; Wu, P.M.; Gali, R.R.; Iyer, L.K.; et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 2012, 122, 3063–3087. [Google Scholar]
- De Felice, B.; Guida, M.; Guida, M.; Coppola, C.; de Mieri, G.; Cotrufo, R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene 2009, 508, 35–40. [Google Scholar]
- Lin, N.; Friedlander, R.M. Regeneration of neuromuscular synapses: Action of microRNA-206. Neurosurgery 2010, 66, N19–N20. [Google Scholar] [CrossRef]
- Fox, M.A.; Sanes, J.R.; Borza, D.B.; Eswarakumar, V.P.; Fässler, R.; Hudson, B.G.; John, S.W.; Ninomiya, Y.; Pedchenko, V.; Pfaff, S.L.; et al. Distinct target-derived signals organize formation, maturation, and maintenance of motor nerve terminals. Cell 2007, 129, 179–193. [Google Scholar] [CrossRef]
- Umemori, H.; Sanes, J.R. Signal regulatory proteins (SIRPS) are secreted presynaptic organizing molecules. J. Biol. Chem. 2008, 283, 34053–34061. [Google Scholar] [CrossRef]
- Beer, H.D.; Bittner, M.; Niklaus, G.; Munding, C.; Max, N.; Goppelt, A.; Werner, S. The fibroblast growth factor binding protein is a novel interaction partner of FGF-7, FGF-10 and FGF-22 and regulates FGF activity: Implications for epithelial repair. Oncogene 2005, 24, 5269–5277. [Google Scholar] [CrossRef]
- Santambrogio, S.; Ricca, A.; Maderna, C.; Ieraci, A.; Aureli, M.; Sonnino, S.; Kulik, W.; Aimar, P.; Bonfanti, L.; Martino, S.; et al. The galactocerebrosidase enzyme contributes to maintain a functional neurogenic niche during early post-natal CNS development. Hum. Mol. Genet. 2012, 21, 4732–4750. [Google Scholar] [CrossRef]
- Neri, M.; Ricca, A.; di Girolamo, I.; Alcala’-Franco, B.; Cavazzin, C.; Orlacchio, A.; Martino, S.; Naldini, L.; Gritti, A. Neural stem cell gene therapy ameliorates pathology and function in a mouse model of globoid cell leukodystrophy. Stem Cells 2011, 29, 1559–1571. [Google Scholar] [CrossRef]
- Lattanzi, A.; Neri, M.; Maderna, C.; di Girolamo, I.; Martino, S.; Orlacchio, A.; Amendola, M.; Naldini, L.; Gritti, A. Widespread enzymatic correction of CNS tissues by a single intracerebral injection of therapeutic lentiviral vector in leukodystrophy mouse models. Hum. Mol. Genet. 2010, 19, 2208–2227. [Google Scholar] [CrossRef]
- Martino, S.; Consiglio, A.; Cavalieri, C.; Tiribuzi, R.; Costanzi, E.; Severini, G.M.; Emiliani, C.; Bordignon, C.; Orlacchio, A. Expression and purification of a human, soluble Arylsulfatase A for Metachromatic Leukodystrophy enzyme replacement therapy. J. Biotechnol. 2005, 117, 243–251. [Google Scholar]
- Orchard, P.J.; Wagner, J.E. Leukodystrophy and gene therapy with a dimmer switch. N. Engl. J. Med. 2011, 364, 572–573. [Google Scholar] [CrossRef]
- Martino, S.; Tiribuzi, R.; Tortori, A.; Conti, D.; Visigalli, I.; Lattanzi, A.; Biffi, A.; Gritti, A.; Orlacchio, A. Specific determination of beta-galactocerebrosidase activity via competitive inhibition of beta-galactosidase. Clin. Chem. 2009, 55, 541–548. [Google Scholar] [CrossRef]
- Visigalli, I.; Ungari, S.; Martino, S.; Park, H.; Cesani, M.; Gentner, B.; Sergi Sergi, L.; Orlacchio, A.; Naldini, L.; Biffi, A. The galactocerebrosidase enzyme contributes to the maintenance of a functional hematopoietic stem cell niche. Blood 2010, 116, 1857–1866. [Google Scholar] [CrossRef]
- Osborn, M.J.; McElmurry, R.T.; Lees, C.J.; DeFeo, A.P.; Chen, Z.Y.; Kay, M.A.; Naldini, L.; Freeman, G.; Tolar, J.; Blazar, B.R. Minicircle DNA-based gene therapy coupled with immune modulation permits long-term expression of α-L-iduronidase in mice with mucopolysaccharidosis type I. Mol. Ther. 2011, 19, 450–460. [Google Scholar]
- Liu, N.; Landreh, M.; Cao, K.; Abe, M.; Hendriks, G.J.; Kennerdell, J.R.; Zhu, Y.; Wang, L.S.; Bonini, N. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 2012, 482, 519–523. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bicchi, I.; Morena, F.; Montesano, S.; Polidoro, M.; Martino, S. MicroRNAs and Molecular Mechanisms of Neurodegeneration. Genes 2013, 4, 244-263. https://doi.org/10.3390/genes4020244
Bicchi I, Morena F, Montesano S, Polidoro M, Martino S. MicroRNAs and Molecular Mechanisms of Neurodegeneration. Genes. 2013; 4(2):244-263. https://doi.org/10.3390/genes4020244
Chicago/Turabian StyleBicchi, Ilaria, Francesco Morena, Simona Montesano, Mario Polidoro, and Sabata Martino. 2013. "MicroRNAs and Molecular Mechanisms of Neurodegeneration" Genes 4, no. 2: 244-263. https://doi.org/10.3390/genes4020244