H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species
Abstract
:1. Introduction
2. H-NS and Its Role in the Regulatory Cascade Controlling the Transcription of Virulence Genes in Shigella
2.1. The H-NS Protein and Its Interactions with DNA
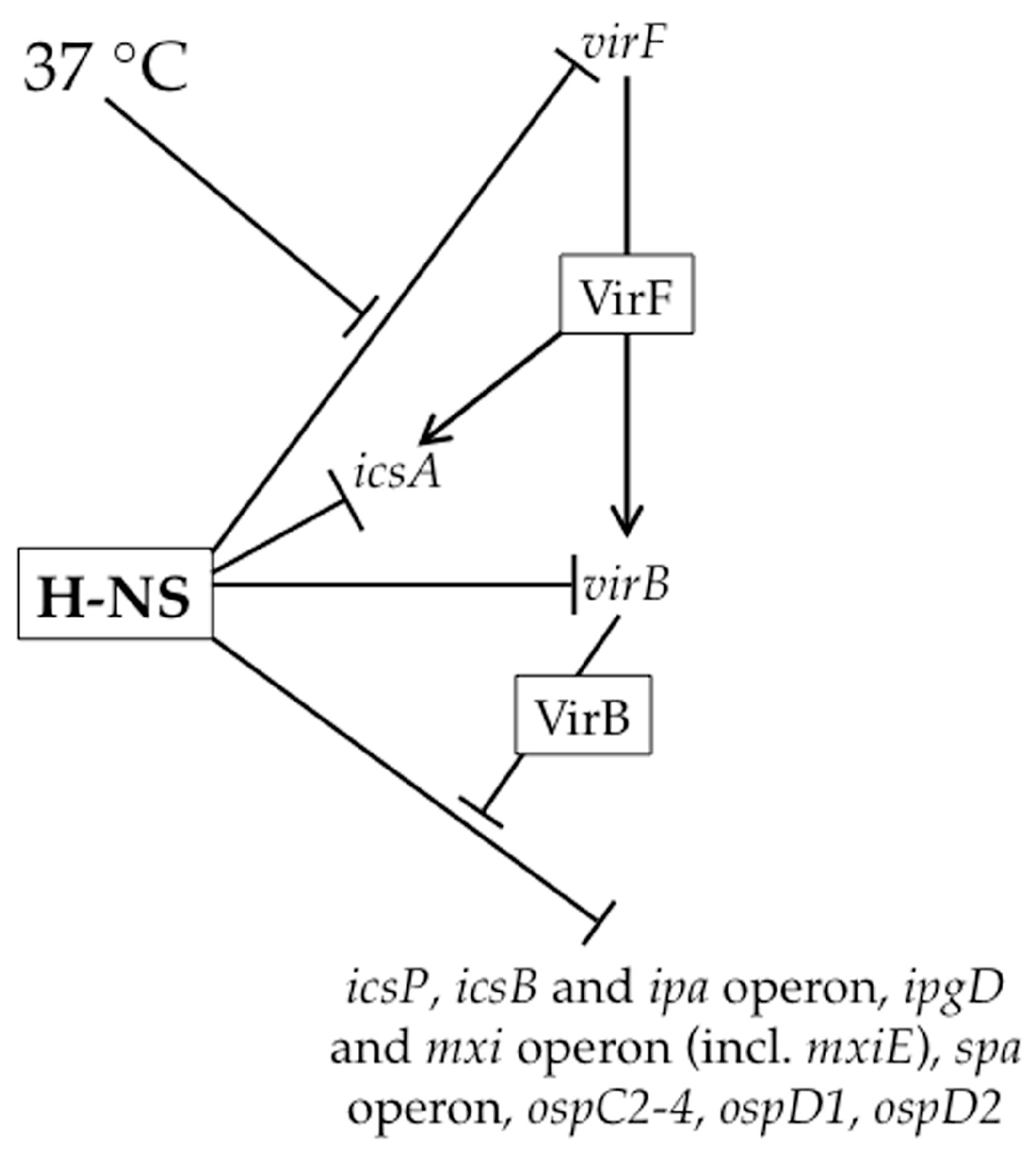
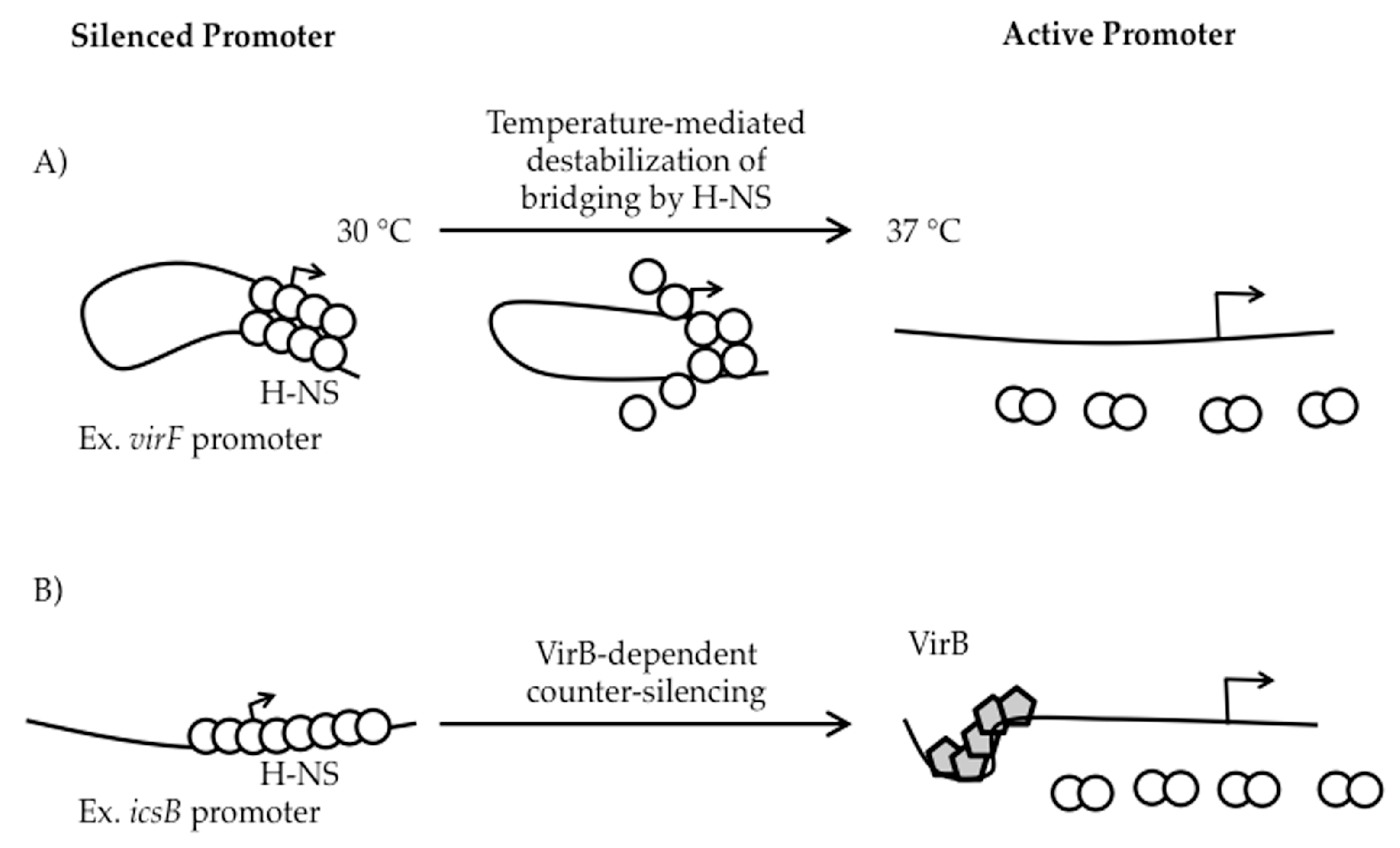
2.2. The Role of H-NS in Shigella Virulence Gene Regulation
3. Activities of the H-NS Paralogues StpA and Sfh and Their Possible Roles in Virulence Gene Regulation
3.1. Reported Activities of StpA and Sfh in E. coli and Salmonella
3.2. Important Considerations When Studying Virulence Gene Regulation by the H-NS Family
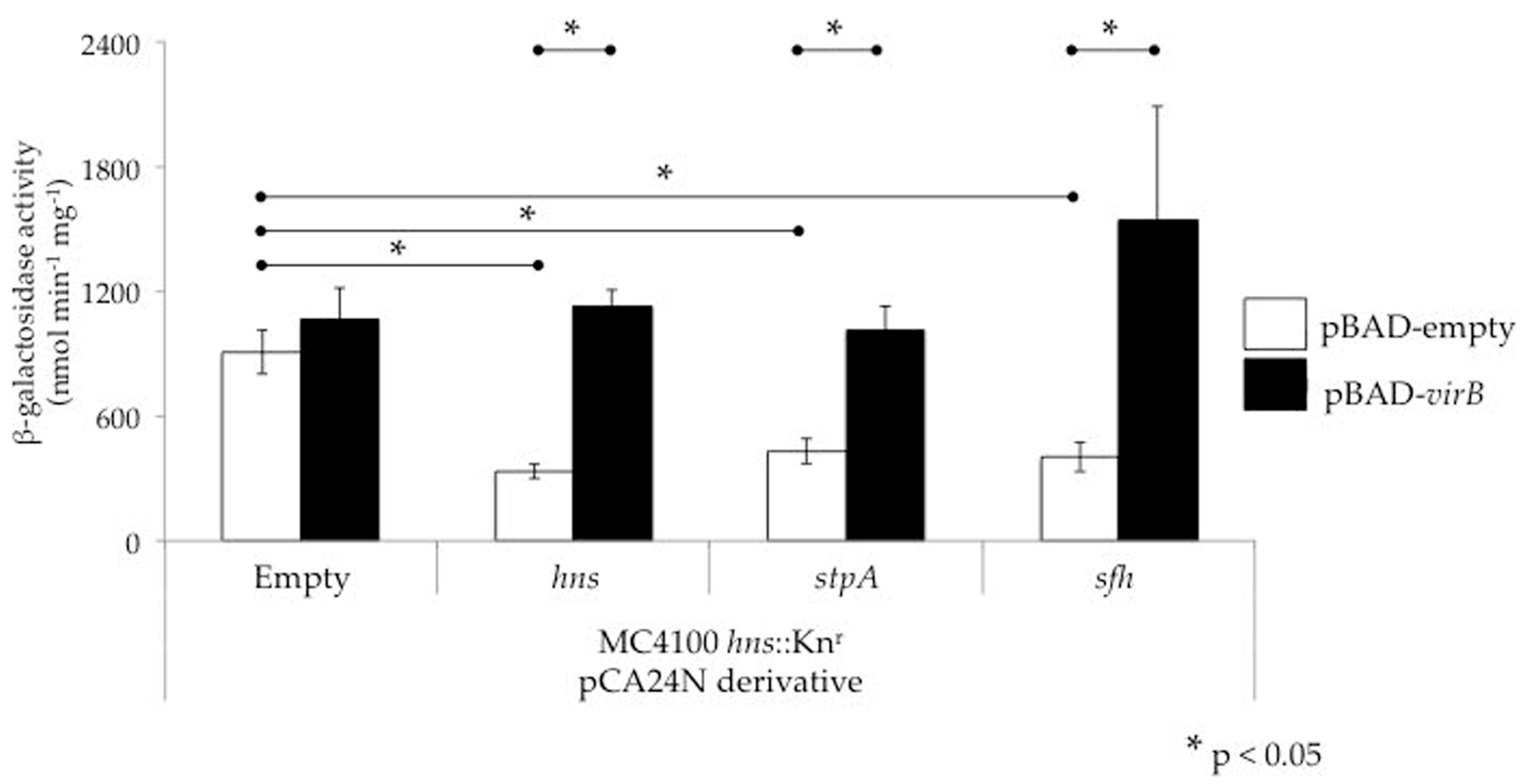
3.3. The Role of StpA and Sfh in Shigella Virulence Gene Regulation
4. Can VirB Alleviate StpA- and Sfh-Mediated Silencing?
5. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Lamberti, L.M.; Bourgeois, A.L.; Fischer Walker, C.L.; Black, R.E.; Sack, D. Estimating Diarrheal Illness and Deaths Attributable to Shigellae and Enterotoxigenic Escherichia coli among Older Children, Adolescents, and Adults in South Asia and Africa. PLoS Negl. Trop. Dis. 2014, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Wang, H.; Lozano, R.; Davis, A.; Liang, X.; Zhou, M. GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar]
- Lan, R.; Lumb, B.; Ryan, D.; Reeves, P.R. Molecular Evolution of Large Virulence Plasmid in Shigella Clones and Enteroinvasive Escherichia coli. Infect. Immun. 2001, 69, 6303–6309. [Google Scholar] [PubMed]
- Venkatesan, M.M.; Goldberg, M.B.; Rose, D.J.; Grotbeck, E.J.; Burland, V.; Blattner, F.R. Complete DNA Sequence and Analysis of the Large Virulence Plasmid of Shigella flexneri. Infect. Immun. 2001, 69, 3271–3285. [Google Scholar] [PubMed]
- Marteyn, B.S.; Gazi, A.D.; Sansonetti, P.J. Shigella: A model of virulence regulation in vivo. Gut Microbes 2012, 3, 104–120. [Google Scholar] [PubMed]
- Dorman, C.J. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 2014, 75, 1–11. [Google Scholar] [PubMed]
- Will, R.W.; Navarre, W.W.; Fang, F.C.; Will, W.R.; Navarre, W.W.; Fang, F.C. Integrated circuits: How transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr. Opin. Microbiol. 2015, 23, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Hommais, F.; Krin, E.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.-P.; Danchin, A.; Bertin, P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; McClelland, M.; Libby, S.J.; Fang, F.C. Silencing of xenogenic DNA by H-NS—Facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007, 21, 1456–1471. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Xia, B.; Liu, J.; Navarre, W.W. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 2012, 15, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Soo, J.; Rao, C.; Leung, A.S.; Ngai, D.H.M.; Ensminger, A.W.; Navarre, W.W. Silencing by H-NS Potentiated the Evolution of Salmonella. PLoS Pathog. 2014, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Stoebel, D.M.; Free, A.; Dorman, C.J. Anti-silencing: Overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 2008, 154, 2533–2545. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J.; Porter, M.E. The Shigella virulence gene regulatory cascade: A paradigm of bacterial gene control mechanisms. Mol. Microbiol. 1998, 29, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Deighan, P.; Beloin, C.; Dorman, C.J. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol. Microbiol. 2003, 48, 1401–1416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Rimsky, S.; Reaban, M.E.; Buc, H.; Belfort, M. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996, 15, 1340–1349. [Google Scholar] [PubMed]
- Doyle, M.; Dorman, C.J. Reciprocal transcriptional and posttranscriptional growth-phase-dependent expression of sfh, a gene that encodes a paralogue of the nucleoid-associated protein H-NS. J. Bacteriol. 2006, 188, 7581–7591. [Google Scholar] [CrossRef] [PubMed]
- Spassky, A.; Rimsky, S.; Garreau, H.; Buc, H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984, 12, 5321–5340. [Google Scholar] [CrossRef] [PubMed]
- Ceschini, S.; Lupidi, G.; Coletta, M.; Pon, C.L.; Fioretti, E.; Angeletti, M. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J. Biol. Chem. 2000, 275, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Rimsky, S.; Buc, H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996, 178, 4335–4343. [Google Scholar] [CrossRef] [PubMed]
- Arold, S.T.; Leonard, P.G.; Parkinson, G.N.; Ladbury, J.E. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. USA 2010, 107, 15728–15732. [Google Scholar] [CrossRef] [PubMed]
- Renault, M.; García, J.; Cordeiro, T.N.; Baldus, M.; Pons, M. Protein oligomers studied by solid-state NMR—The case of the full-length nucleoid-associated protein histone-like nucleoid structuring protein. FEBS J. 2013, 280, 2916–2928. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J.; Hinton, J.C.; Free, A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999, 7, 124–128. [Google Scholar] [CrossRef]
- Bouffartigues, E.; Buckle, M.; Badaut, C.; Travers, A.; Rimsky, S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007, 14, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.; Blot, N.; Bouffartigues, E.; Buckle, M.; Geertz, M.; Gualerzi, C.O.; Mavathur, R.; Muskhelishvili, G.; Pon, C.L.; Rimsky, S.; et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007, 35, 6330–6337. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.R.G.; Li, Y.; Cote, A.; Weirauch, M.T.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Xia, B.; Liu, J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 10690–10695. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Privé, G.G.; Goodsell, D.S.; Dickerson, R.E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc. Natl. Acad. Sci. USA 1988, 85, 6332–6336. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Rimsky, S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008, 11, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Wyman, C.; Goosen, N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000, 28, 3504–3510. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Luijsterburg, M.S.; Krin, E.; Bertin, P.N.; Wagner, R.; Wuite, G.J.L. DNA bridging: A property shared among H-NS-like proteins. J. Bacteriol. 2005, 187, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Noom, M.C.; Wuite, G.J.L. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 2006, 444, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Falconi, M.; Colonna, B.; Prosseda, G.; Micheli, G.; Gualerzi, C.O. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998, 17, 7033–7043. [Google Scholar] [CrossRef] [PubMed]
- Tobe, T.; Yoshikawa, M.; Mizuno, T.; Sasakawa, C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: Activation by VirF and repression by H-NS. J. Bacteriol. 1993, 175, 6142–6149. [Google Scholar] [CrossRef] [PubMed]
- Turner, E.C.; Dorman, C.J. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J. Bacteriol. 2007, 189, 3403–3413. [Google Scholar] [CrossRef] [PubMed]
- Maurelli, A.T.; Blackmon, B.; Curtiss, R., III. Temperature-Dependent Expression of Virulence Genes in Shigella species. Infect. Immun. 1984, 43, 195–201. [Google Scholar] [PubMed]
- Maurelli, A.T.; Sansonetti, P.J. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. USA 1988, 85, 2820–2824. [Google Scholar] [CrossRef] [PubMed]
- Hromockyj, A.E.; Tucker, S.C.; Maurelli, A.T. Temperature regulation of Shigella virulence: Identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA 1 Tyr). Mol. Microbiol. 1992, 6, 2113–2124. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.E.; Dorman, C.J. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 1994, 176, 4187–4191. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.E.; Dorman, C.J. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 1997, 256, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Prosseda, G.; Falconi, M.; Giangrossi, M.; Gualerzi, C.O.; Micheli, G.; Colonna, B. The virF promoter in Shigella: More than just a curved DNA stretch. Mol. Microbiol. 2004, 51, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Ulissi, U.; Fabbretti, A.; Sette, M.; Giuliodori, A.M.; Spurio, R. Time-resolved assembly of a nucleoprotein complex between Shigella flexneri virF promoter and its transcriptional repressor H-NS. Nucleic Acids Res. 2014, 42, 13039–13050. [Google Scholar] [CrossRef] [PubMed]
- Falconi, M.; Prosseda, G.; Giangrossi, M.; Beghetto, E.; Colonna, B. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 2001, 42, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Sasakawa, C.; Yoshikawa, M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol. Microbiol. 1988, 2, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, M.L.; Mounier, J.; D′Hauteville, H.; Coquis-Rondon, M.; Sansonetti, P.J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 1989, 86, 3867–3871. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.B. Actin-Based Motility of Intracellular Microbial Pathogens. Microbiol. Mol. Biol. Rev. 2001, 65, 595–626. [Google Scholar] [CrossRef] [PubMed]
- Tobe, T.; Nagai, S.; Okada, N.; Adler, B.; Yoshikawa, M.; Sasakawa, C. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 1991, 5, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.N.; Giangrossi, M.; Prosseda, G.; Brandi, A.; Di Martino, M.L.; Colonna, B.; Falconi, M. A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 2011, 39, 8122–8134. [Google Scholar] [CrossRef] [PubMed]
- Uchiya, K.; Tobe, T.; Komatsu, K.; Suzuki, T.; Watarai, M.; Fukuda, I.; Yoshikawa, M.; Sasakawa, C. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol. Microbiol. 1995, 17, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Santapaola, D.; Casalino, M.; Petrucca, A.; Presutti, C.; Zagaglia, C.; Berlutti, F.; Colonna, B.; Nicoletti, M. Enteroinvasive Escherichia coli virulence-plasmid-carried apyrase (apy) and ospB genes are organized as a bicistronic operon and are subject to differential expression. Microbiology 2002, 148, 2519–2529. [Google Scholar] [CrossRef] [PubMed]
- Wing, H.J.; Yan, A.W.; Goldman, S.R.; Goldberg, M.B. Regulation of IcsP, the Outer Membrane Protease of the Shigella Actin Tail Assembly Protein IcsA, by Virulence Plasmid Regulators VirF and VirB. J. Bacteriol. 2004, 186, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, T.; Mavris, M.; Martino, M.C.; Bernardini, M.L.; Denamur, E.; Parsot, C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 2005, 151, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, M.I.; Harrison, D.J.; Smith, J.M.; Labahn, S.K.; Levy, K.M.; Wing, H.J. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. J. Bacteriol. 2009, 191, 4047–4050. [Google Scholar] [CrossRef] [PubMed]
- Kane, K.A.; Dorman, C.J. VirB-mediated positive feedback control of the virulence gene regulatory cascade of Shigella flexneri. J. Bacteriol. 2012, 194, 5264–5273. [Google Scholar] [CrossRef] [PubMed]
- Basta, D.W.; Pew, K.L.; Immak, J.A.; Park, H.S.; Picker, M.A.; Wigley, A.F.; Hensley, C.T.; Pearson, J.S.; Hartland, E.L.; Wing, H.J. Characterization of the ospZ promoter in Shigella flexneri and its regulation by VirB and H-NS. J. Bacteriol. 2013, 195, 2562–2572. [Google Scholar] [CrossRef] [PubMed]
- Prosseda, G.; Fradiani, P.A.; Lorenzo, M.; Falconi, M.; Micheli, G.; Casalino, M.; Nicoletti, M.; Colonna, B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 1998, 149, 15–25. [Google Scholar] [CrossRef]
- Gao, X.; Zou, T.; Mu, Z.; Qin, B.; Yang, J.; Waltersperger, S.; Wang, M.; Cui, S.; Jin, Q. Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes. Nucleic Acids Res. 2013, 41, 10529–10541. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Dorman, C.J. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 2003, 47, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; McKenna, S.; Dorman, C.J. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 2002, 277, 15333–15344. [Google Scholar] [CrossRef] [PubMed]
- McKenna, S.; Beloin, C.; Dorman, C.J. In vitro DNA-binding properties of VirB, the Shigella flexneri virulence regulatory protein. FEBS Lett. 2003, 545, 183–187. [Google Scholar] [CrossRef]
- Harrison, D. Transcriptional regulation of the Shigella flexneri icsP Promoter: Silencing and anti-silencing by H-NS and VirB. Ph.D. Thesis, University of Nevada Las Vegas, Las Vegas, NV, USA, August 2010. [Google Scholar]
- Hensley, C.T.; Kamneva, O.K.; Levy, K.M.; Labahn, S.K.; Africa, L.A.; Wing, H.J.; Africa, A.; Wing, H.J. Two promoters and two translation start sites control the expression of the Shigella flexneri outer membrane protease IcsP. Arch. Microbiol. 2011, 193, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Schröder, O.; Wagner, R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 2000, 298, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Kenney, L.J.; Yan, J. Single-molecule studies on the mechanical interplay between DNA supercoiling and H-NS DNA architectural properties. Nucleic Acids Res. 2014, 42, 8369–8378. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Lee, S.Y.; Kenney, L.J.; Yan, J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci. Rep. 2012, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Standish, A.J.; Teh, M.Y.; Tran, E.N.H.; Doyle, M.T.; Baker, P.J.; Morona, R. Unprecedented abundance of protein tyrosine phosphorylation modulates Shigella flexneri virulence. J. Mol. Biol. 2016, 428, 4197–4208. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Deighan, P.; Doyle, M.; Dorman, C.J. Shigella flexneri 2a strain 2457T expresses three members of the H-NS-like protein family: Characterization of the Sfh protein. Mol. Genet. Genomics 2003, 270, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.M.; Schneider, G.; Dobrindt, U.; Emödy, L.; Hacker, J.; Uhlin, B.E. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol. Microbiol. 2010, 75, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Hüttener, M.; Paytubi, S.; Juárez, A. Success in incorporating horizontally transferred genes: The H-NS protein. Trends Microbiol. 2014, 23, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.; Fookes, M.; Ivens, A.; Mangan, M.W.; Wain, J.; Dorman, C.J. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 2007, 315, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Cameron, A.D.; Hokamp, K.; Lucchini, S.; Hinton, J.C.D.; Dorman, C.J. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 2010, 76, 1250–1265. [Google Scholar] [CrossRef] [PubMed]
- Free, A.; Dorman, C.J. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 1997, 179, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Tendeng, C.; Bertin, P.N. H-NS in Gram-negative bacteria: A family of multifaceted proteins. Trends Microbiol. 2003, 11, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.A.; Iwata, A.; Nishimura, A.; Ueda, S.; Ishihama, A. Growth Phase-Dependent Variation in Protein Composition of the Escherichia coli Nucleoid. J. Bac 1999, 181, 6361–6370. [Google Scholar]
- Dame, R.T.; Wyman, C.; Goosen, N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 2001, 83, 231–234. [Google Scholar] [CrossRef]
- Lim, C.J.; Whang, Y.R.; Kenney, L.J.; Yan, J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2012, 40, 3316–3328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Belfort, M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992, 20, 6735. [Google Scholar] [CrossRef] [PubMed]
- Cusick, M.E.; Belfort, M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 1998, 28, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Derbyshire, V.; Salvo, J.L.; Belfort, M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1995, 1, 783–793. [Google Scholar] [PubMed]
- Rajkowitsch, L.; Schroeder, R. Coupling RNA annealing and strand displacement: A FRET-based microplate reader assay for RNA chaperone activity. Biotechniques 2007, 43, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Deighan, P.; Free, A.; Dorman, C.J. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 2000, 38, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Ueguchi, C.; Mizuno, T. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J. Bacteriol. 1996, 178, 3650–3653. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W. H-NS as a Defence System. In Bacterial Chromatin; Dame, R.T., Dorman, C.J., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 251–322. [Google Scholar]
- Yamada, H.; Yoshida, T.; Tanaka, K.; Sasakawa, C.; Mizuno, T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 1991, 230, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Battesti, A.; Tsegaye, Y.M.; Packer, D.G.; Majdalani, N.; Gottesman, S. H-NS Regulation of IraD and IraM antiadaptors for control of RpoS degradation. J. Bacteriol. 2012, 194, 2470–2478. [Google Scholar] [CrossRef] [PubMed]
- Kotlajich, M.V.; Hron, D.R.; Boudreau, B.A.; Sun, Z.; Lyubchenko, Y.L.; Landick, R. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. Elife 2015, 4, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.M.; Chaerkady, R.; Sharma, J.; Díaz-Mejía, J.J.; Tyagi, N.; Renuse, S.; Jacob, H.K.C.; Pinto, S.M.; Sahasrabuddhe, N.A.; Kim, M.-S.; et al. The Escherichia coli Phosphotyrosine Proteome Relates to Core Pathways and Virulence. PLoS Pathog. 2013, 9, e1003403. [Google Scholar] [CrossRef] [PubMed]
- Sondén, B.; Uhlin, B.E. Coordinated and differential expression of histone-like proteins in Escherichia coli: Regulation and function of the H-NS analog StpA. EMBO J. 1996, 15, 4970–4980. [Google Scholar] [PubMed]
- Müller, C.M.; Dobrindt, U.; Nagy, G.; Emödy, L.; Uhlin, B.E.; Hacker, J. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 2006, 188, 5428–5438. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picker, M.A.; Wing, H.J. H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species. Genes 2016, 7, 112. https://doi.org/10.3390/genes7120112
Picker MA, Wing HJ. H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species. Genes. 2016; 7(12):112. https://doi.org/10.3390/genes7120112
Chicago/Turabian StylePicker, Michael A., and Helen J. Wing. 2016. "H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species" Genes 7, no. 12: 112. https://doi.org/10.3390/genes7120112




