Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Control, and Alzheimer’s Disease (AD) Brain Tissues
2.2. Extraction of Total RNA and Protein and Quality Control
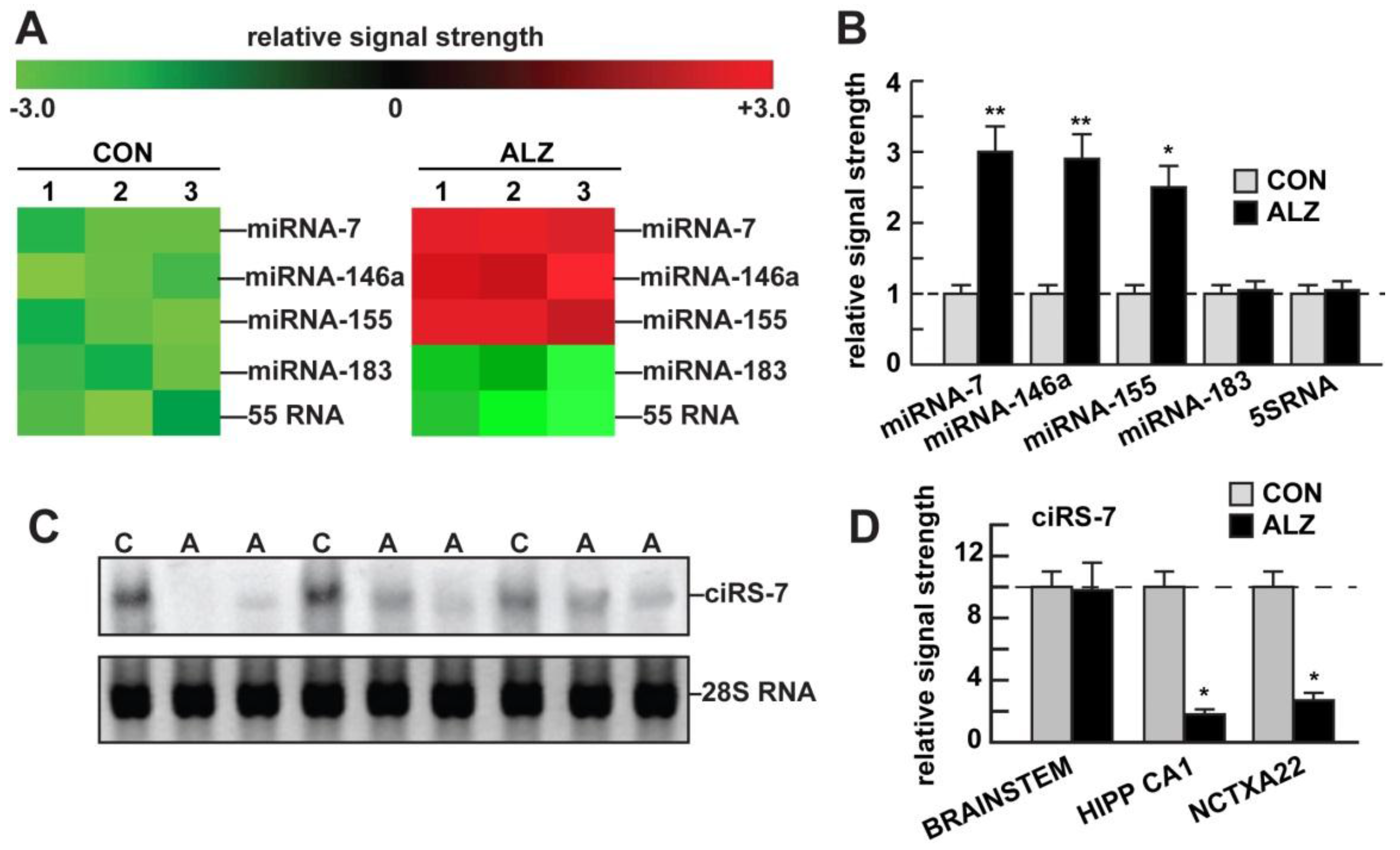
2.3. miRNA Array and LED-Northern Blot Analyses
2.4. Western Blot Analysis of UBE2A and β-actin in AD and Control Tissues
2.5. Statistical Analysis and Data Interpretation
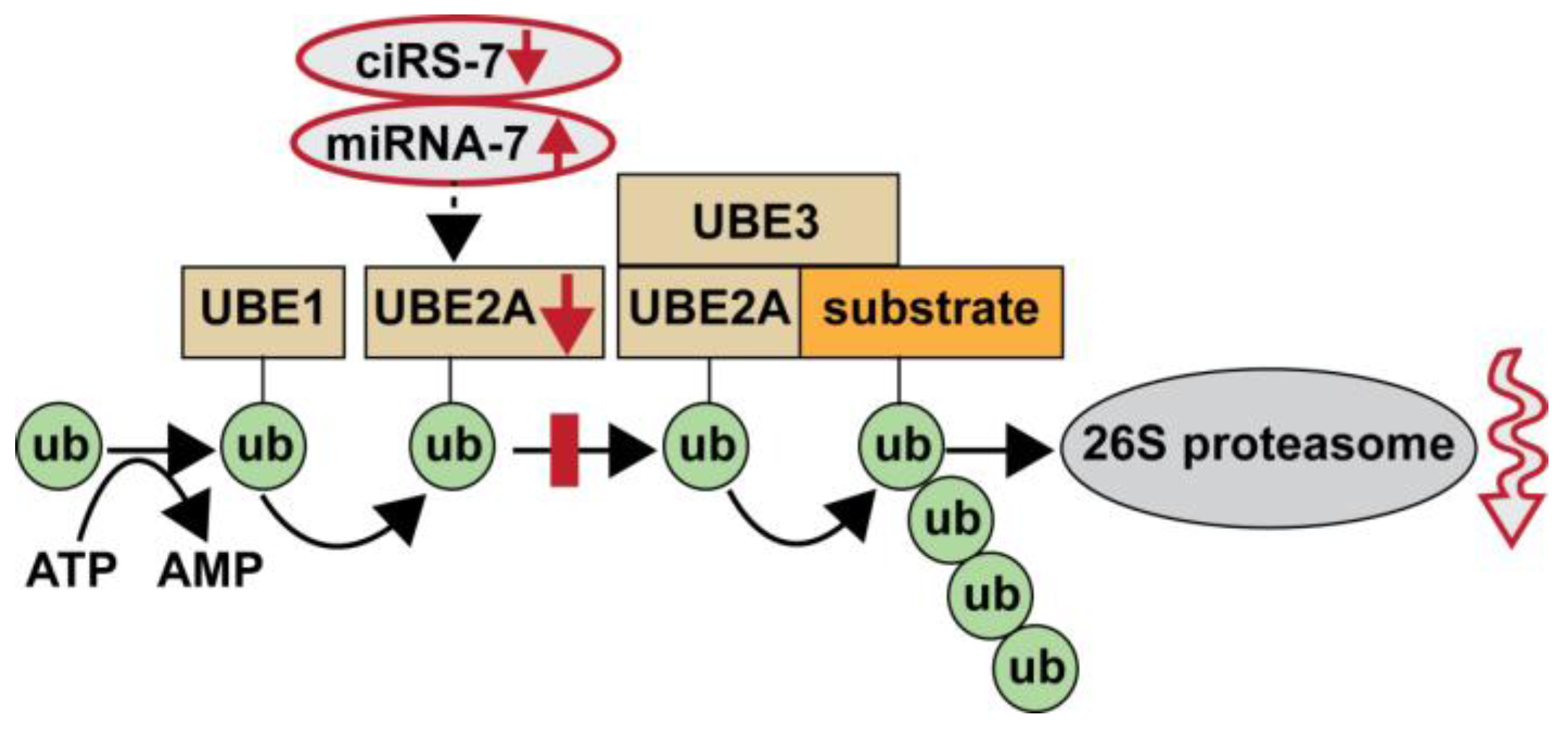
3. Results
4. Discussion
5. Conclusions
Acknowledgements
Conflicts of Interest
References
- Gentier, R.J.; van Leeuwen, F.W. Misframed ubiquitin and impaired protein quality control: An early event in Alzheimer’s disease. Front. Mol. Neurosci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yang, B.; Mo, X.; Xiao, H. Mechanism and regulation of autophagy and its role in neuronal diseases. Mol. Neurobiol. 2015, 52, 1190–1209. [Google Scholar] [CrossRef] [PubMed]
- Suresh, B.; Lee, J.; Kim, K.S.; Ramakrishna, S. The importance of ubiquitination and deubiquitination in cellular reprogramming. Stem Cells Int. 2016, 2016, 6705927. [Google Scholar] [CrossRef] [PubMed]
- McDowel, G.S.; Philpott, A. New insights into the role of ubiquitylation of proteins. Int. Rev. Cell Mol. Biol. 2016, 325, 35–88. [Google Scholar] [CrossRef]
- Ribonuclease R, E. coli. Available online: http://www.epibio.com/docs/default-source/protocols/ribonuclease-r-e-coli.pdf?sfvrsn=8 (accessed on 17 November 2016).
- Nath, D.; Shadan, S. The ubiquitin system. Nature 2009, 458, 421. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Zhao, Y.; Cui, J.G. An NF-kB-sensitive miRNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008, 283, 31315–31322. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Handley, P.; Wong, L.; McLachlan, D.R.C. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem. Res. 1992, 17, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sharp, P.A. A circuitous route to noncoding RNA. Science 2013, 340, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Microcosm targets. Available online: http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/ (accessed on 17 November 2016).
- Zhao, Y.; Pogue, A.I.; Lukiw, W.J. microRNA (miRNA) signaling in the human CNS in sporadic Alzheimer’s disease (AD)—Novel and unique pathological features. Int. J. Mol. Sci. 2015, 16, 30105–30116. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Zhao, Y.; Dua, P.; Rogaev, E.I.; Lukiw, W.J. microRNA-34a-mediated down-Regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS ONE 2016. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Dua, P.; Pogue, A.I.; Eicken, C.; Hill, J.M. Up-regulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J. Toxicol. Environ. Health 2011, 74, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.G.; Li, Y.Y.; Zhao, Y.; Bhattacharjee, S.; Lukiw, W.J. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 2010, 285, 38951–38960. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, P.N.; Dua, P.; Hill, J.M.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 2012, 3, 365–373. [Google Scholar] [PubMed]
- Abe, N.; Hiroshima, M.; Maruyama, H.; Nakashima, Y.; Nakano, Y.; Matsuda, A.; Sako, Y.; Ito, Y.; Abe, H. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew. Chem. Int. Ed. Engl. 2013, 52, 7004–7008. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed]
- van Rossum, D.; Verheijen, B.M.; Pasterkamp, R.J. Circular RNAs: Novel regulators of neuronal development. Front. Mol. Neurosci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Mills, J.D.; Takenaka, K.; Bliim, N.; Halliday, G.M.; Janitz, M. Characterization of circular RNAs landscape in multiple system atrophy brain. J. Neurochem. 2016, 139, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Filippenkov, I.B.; Kalinichenko, E.O.; Limborska, S.A.; Dergunova, L.V. Circular RNAs—One of the enigmas of the brain. Neurogenetics 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Schuman, E. Circular RNAs in brain and other tissues: A functional enigma. Trends Neurosci. 2016, 39, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.L.; Yang, L.; Chen, L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J. Circular RNA Expression: Its potential regulation and function. Trends Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Appel, B. In vitro circularization of RNA. RNA Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Chen, L.L. Characterization of circular RNAs. Methods Mol. Biol. 2016. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Gulyaeva, L.F.; Kushlinskiy, N.E. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cecarini, V.; Bonfili, L.; Amici, M.; Angeletti, M.; Keller, J.N.; Eleuteri, A.M. Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res. 2008. [Google Scholar] [CrossRef] [PubMed]
- Saritas-Yildirim, B.; Silva, E.M. The role of targeted protein degradation in neural development. Genesis 2014, 52, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Horsham, J.L.; Ganda, C.; Kalinowski, F.C.; Brown, R.A.; Epis, M.R.; Leedman, P.J. microRNA-7: A miRNA with expanding roles in development and disease. Int. J. Biochem. Cell Biol. 2015, 69, 215–224. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes 2016, 7, 116. https://doi.org/10.3390/genes7120116
Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes. 2016; 7(12):116. https://doi.org/10.3390/genes7120116
Chicago/Turabian StyleZhao, Yuhai, Peter N. Alexandrov, Vivian Jaber, and Walter J. Lukiw. 2016. "Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7)" Genes 7, no. 12: 116. https://doi.org/10.3390/genes7120116




