Combination of RNA Interference and Stem Cells for Treatment of Central Nervous System Diseases
Abstract
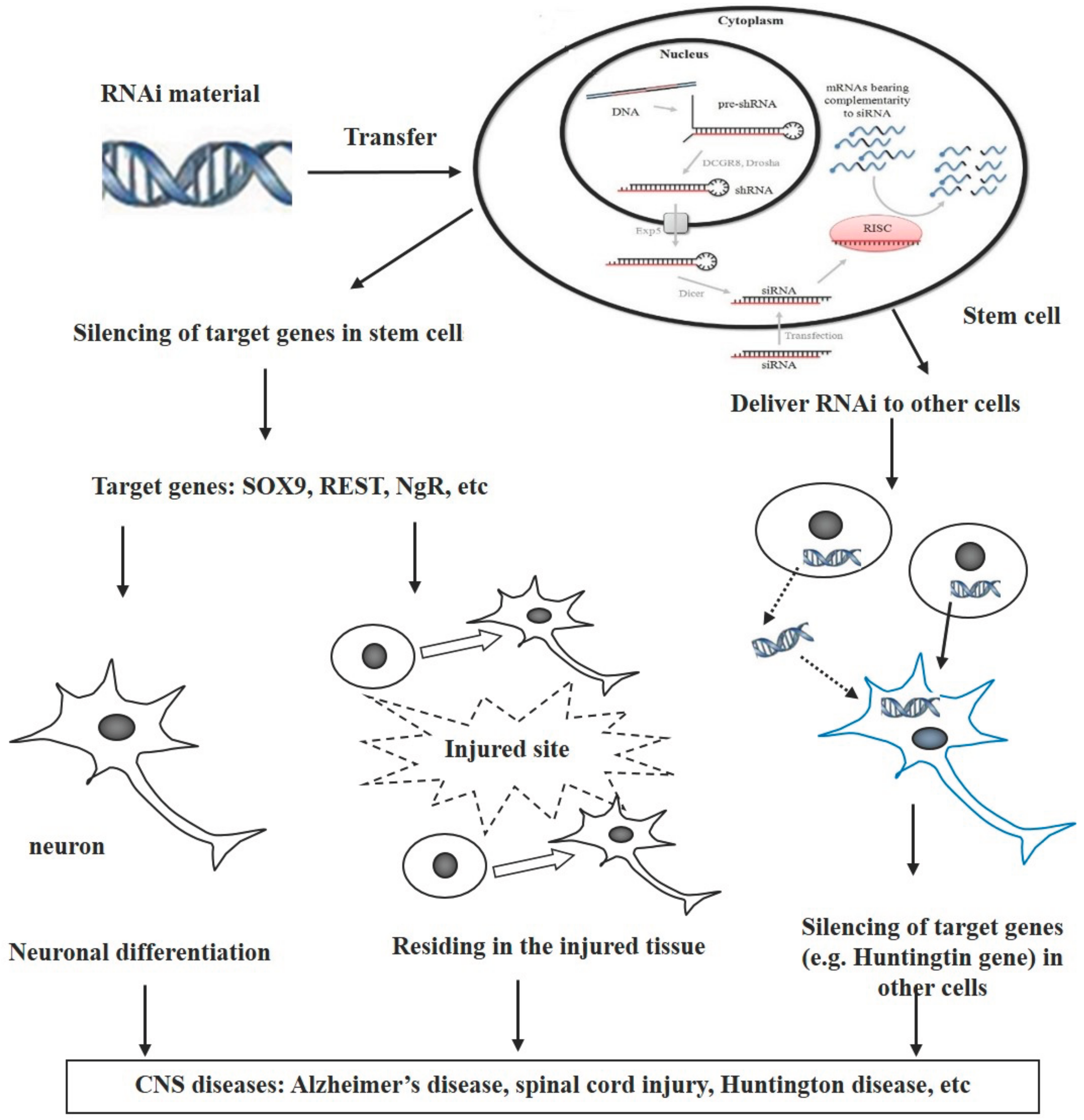
:1. Introduction
2. Endogenous RNAi Pathways in Governing the Neuronal Differentiation and Migration of Stem Cells
3. Combination of RNAi and Stem Cells for Treatment of CNS Diseases
3.1. Alzheimer’s Disease
3.2. Huntington’s Disease
3.3. Spinal Cord Injury
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ager, R.R.; Davis, J.L.; Agazaryan, A.; Benavente, F.; Poon, W.W.; LaFerla, F.M.; Blurton-Jones, M. Human neural stem cells improve cognition and promote synaptic growth in two complementary transgenic models of Alzheimer’s disease and neuronal loss. Hippocampus 2015, 25, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Singh-Bains, M.K.; Tippett, L.J.; Hogg, V.M.; Synek, B.J.; Roxburgh, R.H.; Waldvogel, H.J.; Faull, R.L. Globus pallidus degeneration and clinicopathological features of huntington disease. Ann. Neurol. 2016, 80, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.L.; Nistor, G.; Wyatt, T.; Yin, H.Z.; Poole, A.J.; Weiss, J.H.; Gardener, M.J.; Dijkstra, S.; Fischer, D.F.; Keirstead, H.S. Histological and functional benefit following transplantation of motor neuron progenitors to the injured rat spinal cord. PLoS ONE 2010, 5, e11852. [Google Scholar] [CrossRef] [PubMed]
- Klaric, T.S.; Thomas, P.Q.; Dottori, M.; Leong, W.K.; Koblar, S.A.; Lewis, M.D. A reduction in NPAS4 expression results in delayed neural differentiation of mouse embryonic stem cells. Stem Cell Res. Ther. 2014, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Wang, Z.; Zhao, J.; Gutkind, J.S.; Srivatsan, A.; Zhang, G.; Liao, H.S.; Fu, X.; Jin, A.; et al. Polymeric nanovehicle regulated spatiotemporal real-time imaging of the differentiation dynamics of transplanted neural stem cells after traumatic brain injury. ACS Nano 2015, 9, 6683–6695. [Google Scholar] [CrossRef] [PubMed]
- Guha, P.; Morgan, J.W.; Mostoslavsky, G.; Rodrigues, N.P.; Boyd, A.S. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 2013, 12, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Li, P.; Wang, Y.; Yu, W.; Qin, A.; Liu, M.; Xiang, A.P.; Zhang, W.; Li, W. Nestin regulates neural stem cell migration via controlling the cell contractility. Int. J. Biochem. Cell Biol. 2016, 78, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Kim, Y.J.; Kim, Y.H.; Roh, J.; Kim, E.C.; Lee, H.J.; Kim, S.U.; Yoon, B.W. Long-term effects of magnetically targeted ferumoxide-labeled human neural stem cells in focal cerebral ischemia. Cell Transplant. 2015, 24, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Darsalia, V.; Kallur, T.; Kokaia, Z. Survival, migration and neuronal differentiation of human fetal striatal and cortical neural stem cells grafted in stroke-damaged rat striatum. Eur. J. Neurosci. 2007, 26, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Blurton-Jones, M.; Kitazawa, M.; Martinez-Coria, H.; Castello, N.A.; Muller, F.J.; Loring, J.F.; Yamasaki, T.R.; Poon, W.W.; Green, K.N.; LaFerla, F.M. Neural stem cells improve cognition via bdnf in a transgenic model of Alzheimer disease. Proc. Natl. Acad. Sci. USA 2009, 106, 13594–13599. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Joo, S.S.; Kim, T.K.; Lee, S.H.; Kang, H.; Lee, H.J.; Lim, I.; Matsuo, A.; Tooyama, I.; Kim, Y.B.; et al. Human neural stem cells overexpressing choline acetyltransferase restore cognitive function of kainic acid-induced learning and memory deficit animals. Cell Transplant. 2012, 21, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Zhang, S.Q.; Gu, R.; Liu, J.B.; Li, Y.; Zhu, Q.S. Transplantation of erythropoietin gene-modified neural stem cells improves the repair of injured spinal cord. Neural Regen. Res. 2015, 10, 1483–1490. [Google Scholar] [PubMed]
- Dong, Y.; Yang, L.; Yang, L.; Zhao, H.; Zhang, C.; Wu, D. Transplantation of neurotrophin-3-transfected bone marrow mesenchymal stem cells for the repair of spinal cord injury. Neural Regen. Res. 2014, 9, 1520–1524. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.Q.; Kong, X.H.; Liu, Y.; Ban, D.X.; Ning, G.Z.; Chen, J.T.; Guo, S.F.; Wang, P. Regeneration of spinal cord with cell and gene therapy. Orthop. Surg. 2009, 1, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Low, W.C.; Yau, W.W.; Stanton, L.W.; Marcy, G.; Goh, E.; Chew, S.Y. Directing neuronal differentiation of primary neural progenitor cells by gene knockdown approach. DNA Cell Biol. 2012, 31, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Solanki, A.; Shah, S.; Yin, P.T.; Lee, K.B. Nanotopography-mediated reverse uptake for sirna delivery into neural stem cells to enhance neuronal differentiation. Sci. Rep. 2013, 3, 1553. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.; Lu, P. Therapy of respiratory viral infections with intranasal sirnas. Methods Mol. Biol. 2015, 1218, 251–262. [Google Scholar] [PubMed]
- Olson, S.D.; Kambal, A.; Pollock, K.; Mitchell, G.M.; Stewart, H.; Kalomoiris, S.; Cary, W.; Nacey, C.; Pepper, K.; Nolta, J.A. Examination of mesenchymal stem cell-mediated RNAi transfer to Huntington’s disease affected neuronal cells for reduction of huntingtin. Mol. Cell. Neurosci. 2012, 49, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Murashov, A.K. A brief introduction to RNAi and micrornas in stem cells. Methods Mol. Biol. 2010, 650, 15–25. [Google Scholar] [PubMed]
- Deng, Y.; Wang, C.C.; Choy, K.W.; Du, Q.; Chen, J.; Wang, Q.; Li, L.; Chung, T.K.; Tang, T. Therapeutic potentials of gene silencing by rna interference: Principles, challenges, and new strategies. Gene 2014, 538, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. Rna interference: Mechanisms, technical challenges, and therapeutic opportunities. Methods Mol. Biol. 2015, 1218, 1–15. [Google Scholar] [PubMed]
- Zou, G.M. RNAI in stem cells: Current status and future perspectives. Methods Mol. Biol. 2010, 650, 3–14. [Google Scholar] [PubMed]
- Schultheis, B.; Strumberg, D.; Santel, A.; Vank, C.; Gebhardt, F.; Keil, O.; Lange, C.; Giese, K.; Kaufmann, J.; Khan, M.; et al. First-in-human phase I study of the liposomal RNA interference therapeutic atu027 in patients with advanced solid tumors. J. Clin. Oncol. 2014, 32, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Montanes, J.; Sadaba, B.; Ruz, V.; Gomez-Guiu, A.; Zarranz, J.; Gonzalez, M.V.; Paneda, C.; Jimenez, A.I. Phase I clinical trial of syl040012, a small interfering RNA targeting beta-adrenergic receptor 2, for lowering intraocular pressure. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.; Frank-Kamenetsky, M.; Shulga-Morskaya, S.; Liebow, A.; Bettencourt, B.R.; Sutherland, J.E.; Hutabarat, R.M.; Clausen, V.A.; Karsten, V.; Cehelsky, J.; et al. Effect of an rna interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum ldl cholesterol in healthy volunteers: A randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 2014, 383, 60–68. [Google Scholar] [CrossRef]
- Kakabadze, Z.; Kipshidze, N.; Mardaleishvili, K.; Chutkerashvili, G.; Chelishvili, I.; Harders, A.; Loladze, G.; Shatirishvili, G.; Kipshidze, N.; Chakhunashvili, D.; et al. Phase 1 trial of autologous bone marrow stem cell transplantation in patients with spinal cord injury. Stem Cells Int. 2016, 2016, 6768274. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xie, Z.; Wei, L.; Yang, H.; Yang, S.; Zhu, Z.; Wang, P.; Zhao, C.; Bi, J. Human umbilical cord mesenchymal stem cell-derived neuron-like cells rescue memory deficits and reduce amyloid-beta deposition in an abetapp/ps1 transgenic mouse model. Stem Cell Res. Ther. 2013, 4, 76. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, E.T.; Gustafson, M.P.; Dudakovic, A.; Riester, S.M.; Garces, C.G.; Paradise, C.R.; Takai, H.; Karperien, M.; Cool, S.; Sampen, H.J.; et al. Identification and validation of multiple cell surface markers of clinical-grade adipose-derived mesenchymal stromal cells as novel release criteria for good manufacturing practice-compliant production. Stem Cell Res. Ther. 2016, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.E.; Kim, Y.M.; Jeong, J.S.; Seo, Y.K. The effect of ultrasound for increasing neural differentiation in HBM-MSCS and inducing neurogenesis in ischemic stroke model. Life Sci. 2016, 165, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Turnley, T.; Wishart, A.; Rowe, A.; Kallmeyer, K.; van Vollenstee, F.A.; Thomford, N.E.; Dandara, C.; Chopera, D.; Pepper, M.S.; et al. Fibroblast-derived extracellular matrix induces chondrogenic differentiation in human adipose-derived mesenchymal stromal/stem cells in vitro. Int. J. Mol. Sci. 2016, 17, 1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Lin, S.P.; Hsieh, P.C.; Hung, S.C. Concomitant beige adipocyte differentiation upon induction of mesenchymal stem cells into brown adipocytes. Biochem. Biophys. Res. Commun. 2016, 478, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A microrna array reveals extensive regulation of micrornas during brain development. RNA 2003, 9, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Chen, Y.; Han, Y.; Lv, C.; Tu, G. Overexpression of microrna-124 promotes the neuronal differentiation of bone marrow-derived mesenchymal stem cells. Neural Regen. Res. 2014, 9, 1241–1248. [Google Scholar] [PubMed]
- Gao, Z.; Ding, P.; Hsieh, J. Profiling of rest-dependent micrornas reveals dynamic modes of expression. Front. Neurosci. 2012, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef] [PubMed]
- Laine, S.K.; Alm, J.J.; Virtanen, S.P.; Aro, H.T.; Laitala-Leinonen, T.K. Micrornas mir-96, mir-124, and mir-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J. Cell. Biochem. 2012, 113, 2687–2695. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The microRNA mir-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mrna splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.K. The microrna mir-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Stolt, C.C.; Lommes, P.; Sock, E.; Chaboissier, M.C.; Schedl, A.; Wegner, M. The SOX9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003, 17, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, C. Mesenchymal stem cells deliver exogenous mirnas to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014, 23, 2851–2861. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.; Lee, H.K.; Glasgow, S.M.; Finley, M.; Donti, T.; Gaber, Z.B.; Graham, B.H.; Foster, A.E.; Novitch, B.G.; Gronostajski, R.M.; et al. SOX9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 2012, 74, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Deneen, B.; Ho, R.; Lukaszewicz, A.; Hochstim, C.J.; Gronostajski, R.M.; Anderson, D.J. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 2006, 52, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Tsuyama, J.; Bunt, J.; Richards, L.J.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Shimazaki, T.; Okano, H. MicroRNA-153 regulates the acquisition of gliogenic competence by neural stem cells. Stem Cell Rep. 2015, 5, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, S.M.; Laug, D.; Brawley, V.S.; Zhang, Z.; Corder, A.; Yin, Z.; Wong, S.T.; Li, X.N.; Foster, A.E.; Ahmed, N.; et al. The mir-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J. Neurosci. 2013, 33, 13560–13568. [Google Scholar] [CrossRef] [PubMed]
- Harraz, M.M.; Xu, J.C.; Guiberson, N.; Dawson, T.M.; Dawson, V.L. Mir-223 regulates the differentiation of immature neurons. Mol. Cell. Ther. 2014, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Sun, S.; Li, B.; Huang, C.; Xu, Y.; Han, X.; Xing, Y.; Yan, W. Mir-29a modulates neuronal differentiation through targeting rest in mesenchymal stem cells. PLoS ONE 2014, 9, e97684. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Kan, Q.; Sun, Y.; Wang, S.; Zhang, G.; Peng, T.; Jia, Y. Mir-9 promotes the neural differentiation of mouse bone marrow mesenchymal stem cells via targeting zinc finger protein 521. Neurosci. Lett. 2012, 515, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Bond, H.M.; Mesuraca, M.; Amodio, N.; Mega, T.; Agosti, V.; Fanello, D.; Pelaggi, D.; Bullinger, L.; Grieco, M.; Moore, M.A.; et al. Early hematopoietic zinc finger protein-zinc finger protein 521: A candidate regulator of diverse immature cells. Int. J. Biochem. Cell Biol. 2008, 40, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.; Xie, H.; Zhou, B.; Chia, P.H.; Rizk, P.; Um, M.; Udolph, G.; Yang, H.; Lim, B.; Lodish, H.F. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 2009, 29, 5290–5305. [Google Scholar] [CrossRef] [PubMed]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. Mir-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xiao, Z.; Han, J.; Sun, J.; Ding, W.; Zhao, Y.; Chen, B.; Li, X.; Dai, J. Mir-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting nestin. BMC Neurosci. 2012, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by fmrp-associated micrornas mir-125b and mir-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Han, J.; Xiao, Z.; Chen, T.; Wang, B.; Chen, B.; Liu, S.; Han, S.; Fang, Y.; Wei, J.; et al. The mir-20-rest-wnt signaling axis regulates neural progenitor cell differentiation. Sci. Rep. 2016, 6, 23300. [Google Scholar] [CrossRef] [PubMed]
- Cernilogar, F.M.; Di Giaimo, R.; Rehfeld, F.; Cappello, S.; Lie, D.C. Rna interference machinery-mediated gene regulation in mouse adult neural stem cells. BMC Neurosci. 2015, 16, 60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.Q.; Chen, Z.Y.; Chen, H.M. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene 2016, 593, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.F.; Johnstone, S.E.; Newman, J.J.; Kagey, M.H.; Young, R.A. TCF3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008, 22, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kim, H.J.; Schafer, S.T.; Paquola, A.; Clemenson, G.D.; Toda, T.; Oh, J.; Pankonin, A.R.; Lee, B.S.; Johnston, S.T.; et al. Functional implications of mir-19 in the migration of newborn neurons in the adult brain. Neuron 2016, 91, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Githinji, J.; McLaughlin, B.; Wilczek, K.; Nolta, J. Role of mirnas in neuronal differentiation from human embryonic stem cell-derived neural stem cells. Stem Cell Rev. 2012, 8, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Zhang, R.; Zhang, S.; Shu, Q.; Zhang, D.; Xu, G. Altered expression of micrornas in the neuronal differentiation of human wharton’s jelly mesenchymal stem cells. Neurosci. Lett. 2015, 600, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.C.; Tegge, A.N.; Correa, B.R.; Mahesula, S.; Kohnke, L.Q.; Qiao, M.; Ferreira, M.A.; Kokovay, E.; Penalva, L.O. Mir-124, -128, and -137 orchestrate neural differentiation by acting on overlapping gene sets containing a highly connected transcription factor network. Stem Cells 2016, 34, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Myers, T.J.; Granero-Molto, F.; Longobardi, L.; Li, T.; Yan, Y.; Spagnoli, A. Mesenchymal stem cells at the intersection of cell and gene therapy. Expert Opin. Biol. Ther. 2010, 10, 1663–1679. [Google Scholar] [CrossRef] [PubMed]
- Marei, H.E.; Althani, A.; Suhonen, J.; El Zowalaty, M.E.; Albanna, M.A.; Cenciarelli, C.; Wang, T.; Caceci, T. Common and rare genetic variants associated with Alzheimer’s disease. J. Cell. Physiol. 2016, 231, 1432–1437. [Google Scholar] [CrossRef]
- Xie, Z.; Dong, Y.; Maeda, U.; Xia, W.; Tanzi, R.E. RNA interference silencing of the adaptor molecules SHCC and FE65 differentially affect amyloid precursor protein processing and abeta generation. J. Biol. Chem. 2007, 282, 4318–4325. [Google Scholar] [CrossRef]
- Singer, O.; Marr, R.A.; Rockenstein, E.; Crews, L.; Coufal, N.G.; Gage, F.H.; Verma, I.M.; Masliah, E. Targeting bace1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005, 8, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microrna cluster mir-29a/b-1 in sporadic Alzheimer’s disease correlates with increased bace1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Yang, G.; Song, Y.; Zhou, X.; Deng, Y.; Liu, T.; Weng, G.; Yu, D.; Pan, S. Microrna-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015, 12, 3081–3088. [Google Scholar] [PubMed]
- Pereira, P.A.; Tomás, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant pre-mir-29b for Alzheimer’s disease therapeutics. Sci. Rep. 2016, 6, 19946. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, X.; Wang, L.; Zhang, Y.; Chen, L. Mir-98–5p acts as a target for Alzheimer’s disease by regulating abeta production through modulating SNX6 expression. J. Mol. Neurosci. 2016, 60, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.Y.; Park, H.J.; Kim, H.N.; Oh, S.H.; Bae, J.S.; Ha, H.J.; Lee, P.H. Mesenchymal stem cells enhance autophagy and increase beta-amyloid clearance in Alzheimer disease models. Autophagy 2014, 10, 32–44. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, P.J.; Sha, H.Y.; Ni, J.; Li, M.H.; Gu, G.J. Neural stem cell transplants improve cognitive function without altering amyloid pathology in an app/ps1 double transgenic model of Alzheimer’s disease. Mol. Neurobiol. 2014, 50, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Blurton-Jones, M.; Spencer, B.; Michael, S.; Castello, N.A.; Agazaryan, A.A.; Davis, J.L.; Muller, F.J.; Loring, J.F.; Masliah, E.; LaFerla, F.M. Neural stem cells genetically-modified to express neprilysin reduce pathology in Alzheimer transgenic models. Stem Cell Res. Ther. 2014, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, G.; Lilja, T.; Wallenborg, K.; Falcao, A.M.; Marques, S.C.; Gracias, A.; Solum, D.; Paap, R.; Walfridsson, J.; Teixeira, A.I.; et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Rep. 2014, 3, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Persaud, S.D.; Wei, L.N. Retinoic acid induces ubiquitination-resistant rip140/lsd1 complex to fine-tune pax6 gene in neuronal differentiation. Stem Cells 2016, 34, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cao, H.Q.; Shi, L.Y.; Ng, S.Y.; Stanton, L.W.; Chew, S.Y. Nanofiber topography and sustained biochemical signaling enhance human mesenchymal stem cell neural commitment. Acta Biomater. 2012, 8, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Solanki, A.; Sasmal, P.K.; Lee, K.B. Single vehicular delivery of sirna and small molecules to control stem cell differentiation. J. Am. Chem. Soc. 2013, 135, 15682–15685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Y.; Hu, B.; Lu, Z.; Zhang, J.; Zhang, X. Traceable nanoparticle delivery of small interfering RNA and retinoic acid with temporally release ability to control neural stem cell differentiation for Alzheimer’s disease therapy. Adv. Mater. 2016, 28, 6345–6352. [Google Scholar] [CrossRef] [PubMed]
- Crunkhorn, S. Huntington disease: Boosting ppardelta blocks neurodegeneration. Nat. Rev. Drug Discov. 2016, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Huntington Study Group; Frank, S.; Testa, C.M.; Stamler, D.; Kayson, E.; Davis, C.; Edmondson, M.C.; Kinel, S.; Leavitt, B.; Oakes, D.; et al. Effect of deutetrabenazine on chorea among patients with huntington disease: A randomized clinical trial. JAMA 2016, 316, 40–50. [Google Scholar] [PubMed]
- Paulsen, J.S.; Zhao, H.; Stout, J.C.; Brinkman, R.R.; Guttman, M.; Ross, C.A.; Como, P.; Manning, C.; Hayden, M.R.; Shoulson, I.; et al. Clinical markers of early disease in persons near onset of Huntington’s disease. Neurology 2001, 57, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Conforti, P.; Camnasio, S.; Mutti, C.; Valenza, M.; Thompson, M.; Fossale, E.; Zeitlin, S.; MacDonald, M.E.; Zuccato, C.; Cattaneo, E. Lack of huntingtin promotes neural stem cells differentiation into glial cells while neurons expressing huntingtin with expanded polyglutamine tracts undergo cell death. Neurobiol. Dis. 2013, 50, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.Q.; Staber, P.D.; He, X.; Eliason, S.L.; Martins, I.H.; Mao, Q.; Yang, L.; Kotin, R.M.; Paulson, H.L.; Davidson, B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA 2005, 102, 5820–5825. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, R.L.; McBride, J.L.; Martins, I.; Shen, S.; Xing, Y.; Carter, B.J.; Davidson, B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. J. Am. Soc. Gene Ther. 2009, 17, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.; Okada, T.; Kurosawa, M.; Oyama, F.; Ozawa, K.; Nukina, N. RAAV-mediated shRNA ameliorated neuropathology in huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006, 343, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Pendergraff, H.; Liu, J.; Kordasiewicz, H.B.; Cleveland, D.W.; Swayze, E.E.; Lima, W.F.; Crooke, S.T.; Prakash, T.P.; Corey, D.R. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 2012, 150, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Miniarikova, J.; Zanella, I.; Huseinovic, A.; van der Zon, T.; Hanemaaijer, E.; Martier, R.; Koornneef, A.; Southwell, A.L.; Hayden, M.R.; van Deventer, S.J.; et al. Design, characterization, and lead selection of therapeutic miRNAs targeting huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids 2016, 5, e297. [Google Scholar] [CrossRef] [PubMed]
- Keeler, A.M.; Sapp, E.; Chase, K.; Sottosanti, E.; Danielson, E.; Pfister, E.; Stoica, L.; DiFiglia, M.; Aronin, N.; Sena-Esteves, M. Cellular analysis of silencing the Huntington’s disease gene using AAV9 mediated delivery of artificial micro RNA into the striatum of q140/q140 mice. J. Huntingt. Dis. 2016, 5, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Hoss, A.G.; Labadorf, A.; Latourelle, J.C.; Kartha, V.K.; Hadzi, T.C.; Gusella, J.F.; MacDonald, M.E.; Chen, J.F.; Akbarian, S.; Weng, Z.; et al. Mir-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genom. 2015, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Kocerha, J.; Xu, Y.; Prucha, M.S.; Zhao, D.; Chan, A.W. MicroRNA-128a dysregulation in transgenic Huntington’s disease monkeys. Mol. Brain 2014, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Parekh, V. Mir-34b-a novel plasma marker for huntington disease? Nat. Rev. Neurol. 2011, 7, 304. [Google Scholar] [CrossRef] [PubMed]
- Kozlowska, E.; Krzyzosiak, W.J.; Koscianska, E. Regulation of huntingtin gene expression by miRNA-137, -214, -148a, and their respective isomirs. Int. J. Mol. Sci. 2013, 14, 16999–17016. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Palliser, D.; Lieberman, J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006, 13, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Fehlings, M.; Singh, A.; Tetreault, L.; Kalsi-Ryan, S.; Nouri, A. Global prevalence and incidence of traumatic spinal cord injury. Clin. Epidemiol. 2014, 6, 309–331. [Google Scholar] [CrossRef] [PubMed]
- Kraft, J.; Karpenko, A.; Rincon, F. Intraoperative targeted temperature management in acute brain and spinal cord injury. Curr. Neurol. Neurosci. Rep. 2016, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- National Spinal Cord Injury Statistical Center. Spinal cord injury facts and figures at a glance. J. Spinal Cord Med. 2011, 34, 620–621. [Google Scholar]
- Ning, B.; Gao, L.; Liu, R.H.; Liu, Y.; Zhang, N.S.; Chen, Z.Y. MicroRNAs in spinal cord injury: Potential roles and therapeutic implications. Int. J. Biol. Sci. 2014, 10, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Theis, T.; Yoo, M.; Park, C.S.; Chen, J.; Kugler, S.; Gibbs, K.M.; Schachner, M. Lentiviral delivery of mir-133b improves functional recovery after spinal cord injury in mice. Mol. Neurobiol. 2016, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Louw, A.M.; Kolar, M.K.; Novikova, L.N.; Kingham, P.J.; Wiberg, M.; Kjems, J.; Novikov, L.N. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Sandner, B.; Prang, P.; Rivera, F.J.; Aigner, L.; Blesch, A.; Weidner, N. Neural stem cells for spinal cord repair. Cell Tissue Res. 2012, 349, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Yu, F.; Hong Yu, Y.; Srivats, H.; Dawe, G.S.; Ahmed, S. Nogor1 and pirb signaling stimulates neural stem cell survival and proliferation. Stem Cells 2014, 32, 1636–1648. [Google Scholar] [CrossRef] [PubMed]
- Rolando, C.; Parolisi, R.; Boda, E.; Schwab, M.E.; Rossi, F.; Buffo, A. Distinct roles of nogo-a and nogo receptor 1 in the homeostatic regulation of adult neural stem cell function and neuroblast migration. J. Neurosci. 2012, 32, 17788–17799. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Cen, J.S.; Wang, J.; Qin, G.; Long, L.; Wang, L.; Wei, F.; Xiang, Q.; Deng, D.Y.; Wan, Y. Targeted inhibition of leucine-rich repeat and immunoglobulin domain-containing protein 1 in transplanted neural stem cells promotes neuronal differentiation and functional recovery in rats subjected to spinal cord injury. Crit. Care Med. 2016, 44, e146–e157. [Google Scholar] [CrossRef] [PubMed]
- Thakore-Shah, K.; Koleilat, T.; Jan, M.; John, A.; Pyle, A.D. REST/NRSF knockdown alters survival, lineage differentiation and signaling in human embryonic stem cells. PLoS ONE 2015, 10, e0145280. [Google Scholar] [CrossRef] [PubMed]
- Low, W.C.; Rujitanaroj, P.O.; Lee, D.K.; Messersmith, P.B.; Stanton, L.W.; Goh, E.; Chew, S.Y. Nanofibrous scaffold-mediated rest knockdown to enhance neuronal differentiation of stem cells. Biomaterials 2013, 34, 3581–3590. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.J.; Smirnov, S.V.; Murthy, R.G.; Rameshwar, P. Synergy between the re-1 silencer of transcription and nfkappab in the repression of the neurotransmitter gene tac1 in human mesenchymal stem cells. J. Biol. Chem. 2007, 282, 30039–30050. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Klassert, T.E.; Greco, S.J.; Patel, S.A.; Munoz, J.L.; Reddy, B.Y.; Bryan, M.; Campbell, N.; Kokorina, N.; Sabaawy, H.E.; et al. Developmental regulation of tac1 in peptidergic-induced human mesenchymal stem cells: Implication for spinal cord injury in zebrafish. Stem Cells Dev. 2012, 21, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Lu, X.M.; Zhu, F.; Huang, P.; Yu, Y.; Zeng, L.; Long, Z.Y.; Wu, Y.M. The use of a gold nanoparticle-based adjuvant to improve the therapeutic efficacy of HNGR-FC protein immunization in spinal cord-injured rats. Biomaterials 2011, 32, 7988–7998. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.P.; Fournier, A.; GrandPre, T.; Strittmatter, S.M. Myelin-associated glycoprotein as a functional ligand for the nogo-66 receptor. Science 2002, 297, 1190–1193. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Koprivica, V.; Kim, J.A.; Sivasankaran, R.; Guo, Y.; Neve, R.L.; He, Z. Oligodendrocyte-myelin glycoprotein is a nogo receptor ligand that inhibits neurite outgrowth. Nature 2002, 417, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Geoffroy, C.G.; Chan, A.F.; Tolentino, K.E.; Crawford, M.J.; Leal, M.A.; Kang, B.; Zheng, B. Assessing spinal axon regeneration and sprouting in nogo-, mag-, and omgp-deficient mice. Neuron 2010, 66, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Zhao, L.; Li, H.; Wang, S.; Shen, Y. Bone marrow mesenchymal stem cells with nogo-66 receptor gene silencing for repair of spinal cord injury. Neural Regen. Res. 2014, 9, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Kim, J.A.; Sivasankaran, R.; Segal, R.; He, Z. P75 interacts with the nogo receptor as a co-receptor for nogo, mag and omgp. Nature 2002, 420, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Lee, X.; Shao, Z.; Thill, G.; Ji, B.; Relton, J.; Levesque, M.; Allaire, N.; Perrin, S.; Sands, B.; et al. Lingo-1 is a component of the nogo-66 receptor/p75 signaling complex. Nat. Neurosci. 2004, 7, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Loov, C.; Fernqvist, M.; Walmsley, A.; Marklund, N.; Erlandsson, A. Neutralization of lingo-1 during in vitro differentiation of neural stem cells results in proliferation of immature neurons. PLoS ONE 2012, 7, e29771. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, Z.; Zheng, S.; Chen, L.; Wan, Y.; Deng, Y.; Yang, R. Lingo-1 shRNA and notch signaling inhibitor dapt promote differentiation of neural stem/progenitor cells into neurons. Brain Res. 2016, 1634, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.F.; Cen, J.S.; Zhong, Q.; Chen, L.; Wang, J.; Deng, D.Y.; Wan, Y. The promotion of functional recovery and nerve regeneration after spinal cord injury by lentiviral vectors encoding lingo-1 shrna delivered by pluronic f-127. Biomaterials 2013, 34, 1686–1700. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kelly, C.E.; Goh, K.Y.; Lim, K.B.; Ibanez, C.F. Death domain signaling by disulfide-linked dimers of the p75 neurotrophin receptor mediates neuronal death in the CNS. J. Neurosci. 2016, 36, 5587–5595. [Google Scholar] [CrossRef] [PubMed]
- Edalat, H.; Hajebrahimi, Z.; Movahedin, M.; Tavallaei, M.; Amiri, S.; Mowla, S.J. P75ntr suppression in rat bone marrow stromal stem cells significantly reduced their rate of apoptosis during neural differentiation. Neurosci. Lett. 2011, 498, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Edalat, H.; Hajebrahimi, Z.; Pirhajati, V.; Movahedin, M.; Tavallaei, M.; Soroush, M.R.; Mowla, S.J. Transplanting p75-suppressed bone marrow stromal cells promotes functional behavior in a rat model of spinal cord injury. Iran. Biomed. J. 2013, 17, 140–145. [Google Scholar] [PubMed]
- Plemel, J.R.; Keough, M.B.; Duncan, G.J.; Sparling, J.S.; Yong, V.W.; Stys, P.K.; Tetzlaff, W. Remyelination after spinal cord injury: Is it a target for repair? Prog. Neurobiol. 2014, 117, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Gottle, P.; Hartung, H.P.; Kury, P. Pushing forward: Remyelination as the new frontier in CNS diseases. Trends Neurosci. 2016, 39, 246–263. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Browning, J.L.; Lee, X.; Scott, M.L.; Shulga-Morskaya, S.; Allaire, N.; Thill, G.; Levesque, M.; Sah, D.; McCoy, J.M.; et al. Taj/troy, an orphan TNF receptor family member, binds nogo-66 receptor 1 and regulates axonal regeneration. Neuron 2005, 45, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Baer, A.S.; Syed, Y.A.; Kang, S.U.; Mitteregger, D.; Vig, R.; Ffrench-Constant, C.; Franklin, R.J.; Altmann, F.; Lubec, G.; Kotter, M.R. Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of FYN-RHOA and protein kinase C signalling. Brain J. Neurol. 2009, 132, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Kotter, M.R.; Li, W.W.; Zhao, C.; Franklin, R.J. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J. Neurosci. 2006, 26, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, S.; Sun, Q.; Li, Z.; Xu, F.; Hou, C.; Harada, T.; Chu, M.; Xu, K.; Feng, X.; et al. Inhibition of troy promotes OPC differentiation and increases therapeutic efficacy of OPC graft for spinal cord injury. Stem Cells Dev. 2014, 23, 2104–2118. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.T.; Kerjan, G.; Bielas, S.L.; Lee, J.E.; Fenstermaker, A.G.; Novarino, G.; Gleeson, J.G. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microrna dysregulation. Neuron 2014, 82, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, X.-Q.; Wang, L.; Wang, F.-G.; Zhao, X.-M.; Zhang, H.-T. Combination of RNA Interference and Stem Cells for Treatment of Central Nervous System Diseases. Genes 2017, 8, 135. https://doi.org/10.3390/genes8050135
Hou X-Q, Wang L, Wang F-G, Zhao X-M, Zhang H-T. Combination of RNA Interference and Stem Cells for Treatment of Central Nervous System Diseases. Genes. 2017; 8(5):135. https://doi.org/10.3390/genes8050135
Chicago/Turabian StyleHou, Xue-Qin, Lei Wang, Fu-Gang Wang, Xiao-Min Zhao, and Han-Ting Zhang. 2017. "Combination of RNA Interference and Stem Cells for Treatment of Central Nervous System Diseases" Genes 8, no. 5: 135. https://doi.org/10.3390/genes8050135




