High Prevalence of Quorum-Sensing and Quorum-Quenching Activity among Cultivable Bacteria and Metagenomic Sequences in the Mediterranean Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Bacterial Quantification, and Strain Isolation
2.2. 16S-Based Bacterial Identification
2.3. Quorum-Sensing Activity Assay
2.4. Quorum-Quenching Activity Assay
2.5. Characterization of AHL-Degradation Activity
2.6. Metagenomic Samples
2.7. Metagenomic Analysis
2.8. Data Availability
2.9. Statistical Analysis
3. Results
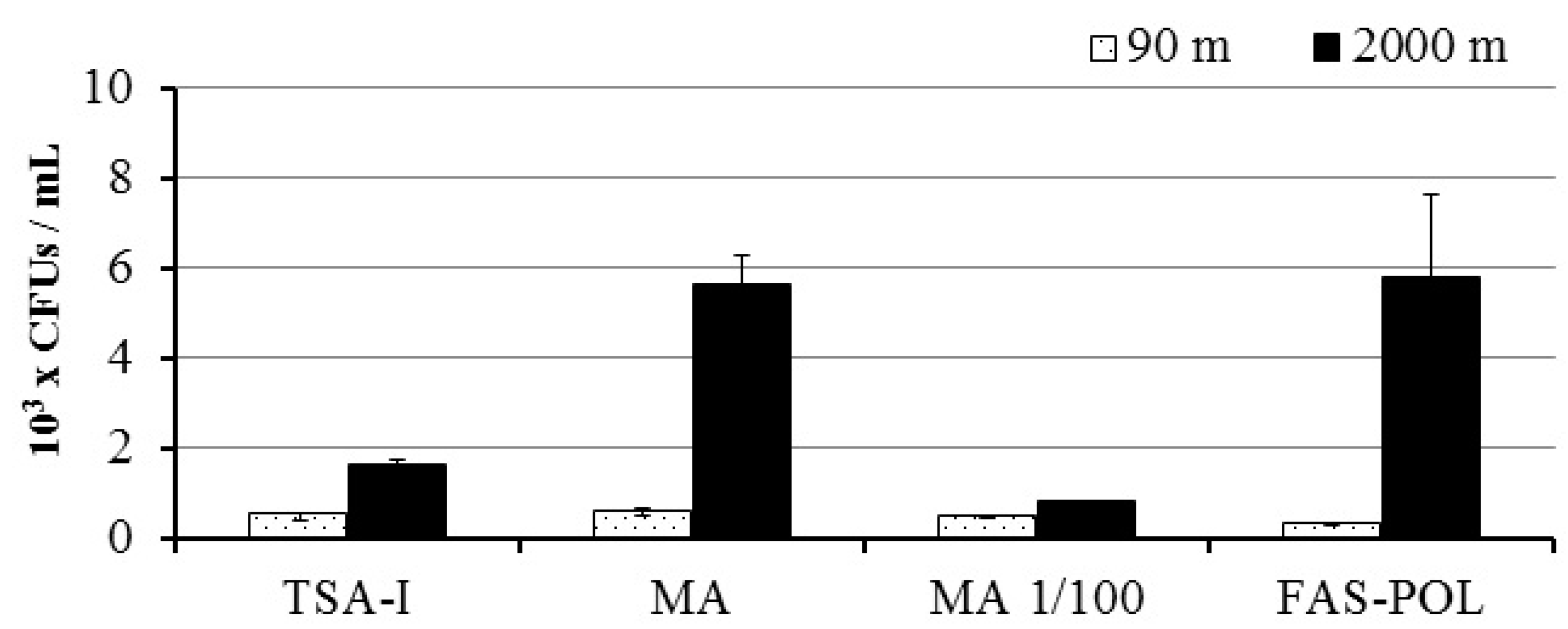
3.1. Bacterial Growth and Isolation
3.2. Taxonomic Diversity of the Cultivable Strains
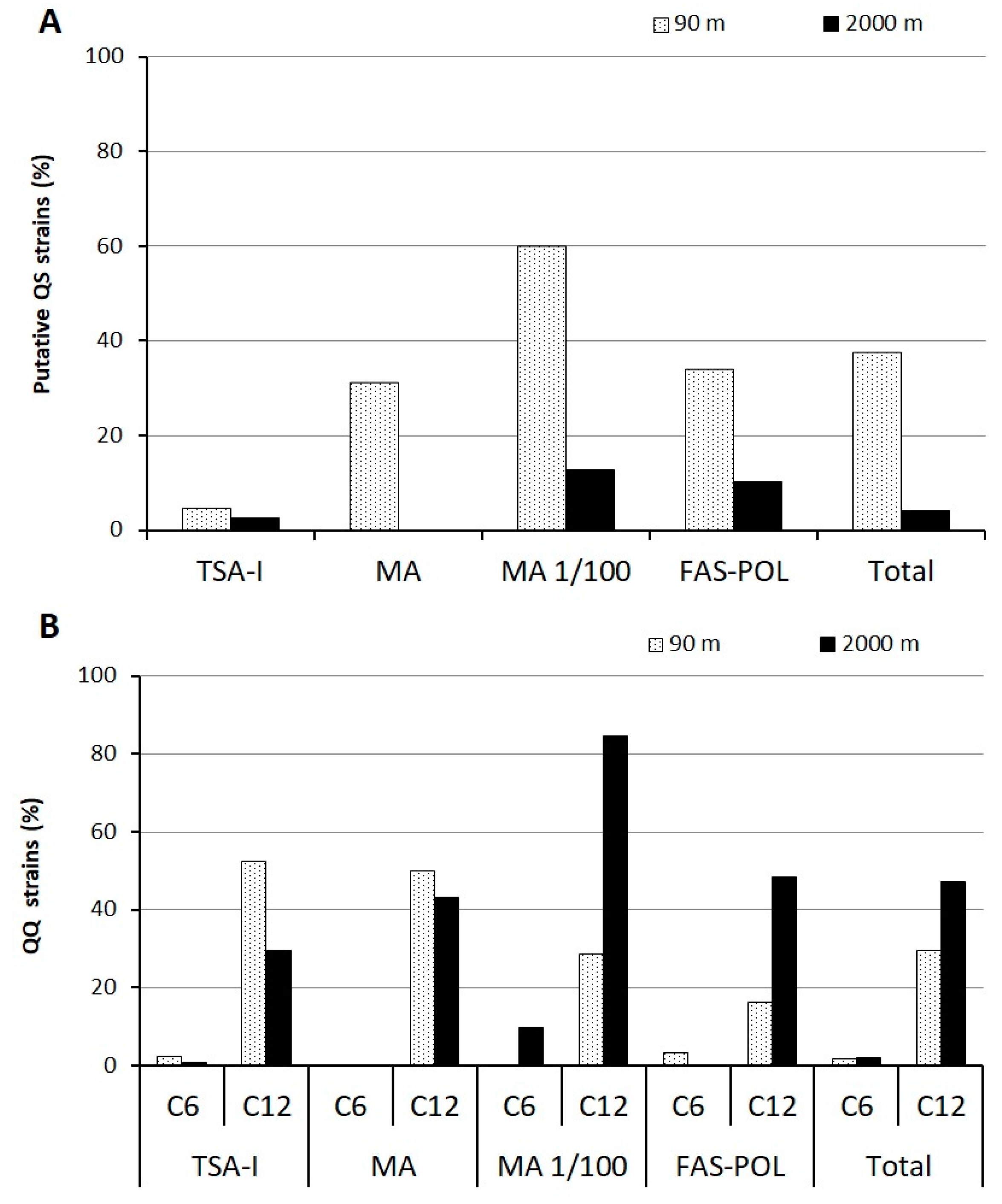
3.3. Quorum-Sensing and Quorum-Quenching Activities among Cultivable Bacteria
3.4. Identification of the Wide-Spectrum Quorum-Quenching Strains and Characterization of Quorum-Quenching Activity
3.5. Quorum-Sensing and Quorum-Quenching Genes in Metagenomic Samples
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fuqua, W.C.; Winans, C.; Greenber, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Dobretsov, S.; Teplitski, M.; Paul, V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling 2009, 5, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannieres, M.; Moréra, S.; Dessaux, Y.; Faure, D.D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. 2015, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Harder, T.; Rice, S.A.; Dobretsov, S.; Thomas, T.; Carré-Mlouka, A.; Kjelleberg, S.; Steinberg, P.; McDougald, D. Bacterial Communication Systems. In Oustanding Marine Molecules: Chemistry, Biology, Analysis; La Barre, S., Kornprobst, J.M., Eds.; Wiley-VCH: Weinheim, Germany, 2014; pp. 173–187. [Google Scholar]
- Biswa, P.; Doble, M. Production of acylated homoserine lactone by Gram-Positive bacteria isolated from marine water. FEMS Microbiol. Lett. 2013, 343, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Gilson, L.; Kuo, A.; Dunlap, P.V. AinS and a new family of Autoinducer synthesis proteins. J. Bacteriol. 1995, 177, 6946–6951. [Google Scholar] [CrossRef] [PubMed]
- Laue, B.E.; Jiang, Y.; Chhabra, S.R.; Jacob, S.; Stewart, G.S.A.; Hardam, A.; Downie, J.A.; O’Gara, F.; Williams, P. The biocontrol strain Pseudomonas fluorescens F113 produces the rhizobium small bacteriocin, N-(3-Hydroxy-7-Cis-Tetradecenoyl) Homoserine Lactone, via HdtS, a putative novel N-acylhomoserine lactone synhtase. Microbiology 2000, 146, 2469–2480. [Google Scholar] [CrossRef] [PubMed]
- Patankar, A.V.; González, J.E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 2009, 33, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Bode, H.B.; Heermann, R. Languages and dialects: Bacterial communication beyond homoserine lactones. Trends Microbiol. 2015, 23, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Cude, W.; Buchan, A. Acyl homoserine lactone-baseed quorum sensing in the Roseobacter clade:complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cicirelli, E.M.; Williamson, H.; Tait, K.; Fuqua, C. Acylated homoserine lactone signalling in marine bacterial systems. In Chemical Communication among Bacteria; Winans, S.C., Bassler, B.L., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 251–271. [Google Scholar]
- Hmelo, L.R. Quorum sensing in marine microbial environments. Annu. Rev. Mar. Sci. 2017, 9, 257–281. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Grossart, H.P.; Schlingloff, A.; Kiorboe, T. Possible quorum sensing in marine snow bacteria: Produtcion of acylated homoserine lactones by Roseobacter strains. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Shupp, P.J.; Baillie, H.J.; Charlton, T.S.; De Nys, R.; Kjelleberg, S.; Steinberg, P.D. Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 2004, 70, 4387–4389. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Döbler, I.; Thiel, V.; Eberl, L.; Allgaier, M.; Bodor, A.; Meyer, S.; Ebner, S.; Henning, A.; Pukall, R.; Schulz, S. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chem. Biochem. 2005, 6, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Avendaño-Herrera, R.; Magariños, B.; Cámara, M.; Otero, A. Acylhomoserine-lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga–Flavobacterium–Bacteroides (CFB) group. FEMS Microbiol. Lett. 2010, 304, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Jatt, A.N.; Tang, K.; Liu, J.; Zhang, Z.; Zhang, X.H. Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea anatatis. FEMS Microbiol. Ecol. 2015, 91, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rolland, J.L.; Stien, D.; Sanchez-Ferandin, S.; Lami, E. Quorum sensing and quorum quenching in the phycosphere of phytoplankton: A case of chemical interactions ecology. J. Chem. Ecol. 2016, 42, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Ki, J.S.; Case, R.J.; Qian, P.Y. Diversity and acyl-homoserine lactone production among subtidal biofilm-forming bacteria. Aquat. Microb. Ecol. 2008, 52, 185–193. [Google Scholar] [CrossRef]
- Mohamed, N.M.; Cicirelli, E.M.; Kan, J.; Chen, F.; Fuqua, C.; Hill, R.T. Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 2008, 10, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Tait, K.; Hutchinson, Z.; Thompson, F.L.; Munn, C.B. Quorum sensing signal production and inhibition by coral-associated Vibrios. Environ. Microbiol. Rep. 2010, 2, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Ransome, E.; Munn, C.B.; Halliday, N.; Cámara, M.; Tait, K. Diverse profiles of N-acyl-homoserine lactone molecules found in cnidarians. FEMS Microbiol. Ecol. 2014, 87, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Hmelo, L.R.; Tracy, J.M.; Van Mooy, B.A.S. Possible influence of bacterial quorum sensing on the hydrolisys of sinking particulate organic carbon in marine environments. Environ. Microbiol. Rep. 2011, 3, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Joint, I.; Tait, K.; Callow, M.E.; Milton, D.; Williams, P.; Cámara, M. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 2002, 298, 1207. [Google Scholar] [CrossRef] [PubMed]
- Tait, K.; Joint, I.; Daykin, M.; Milton, D.L.; Williams, P.; Cámara, M. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 2005, 7, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Goecke, F.; Labes, A.; Wiese, J.; Imhoff, J.F. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 2012, 409, 267–300. [Google Scholar] [CrossRef]
- Van Mooy, B.; Hmelo, L.; Sofe, L.E.; Campagna, S.R.; Mau, A.L.; Dhyrman, S.T.; Heithoff, A.; Webb, E.; Momper, L.; Mincer, T.J. Quorum sensing control of phosphorous acquisition in Trichodesmium consortia. ISME J. 2012, 6, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.; Goecke, F.; Labes, A.; Sobretsov, S.; Weinberg, F. The second skin: Ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 2012, 3, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Alternatives to antibiotics to control bacteria infections: Luminescent vibriosis in aquaculture as an example. Trends Biotechnol. 2007, 25, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.; Visscher, P.T.; Ferry, J.; Kawaguchi, T.; He, L.; Przqkop, K.M.; Norman, R.S.; Reid, R.P. Autoinducers extracted from microbial mats reveals a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may reltae to diel pH. Environ. Microbiol. 2009, 11, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Doberva, M.; Sanchez-Ferandin, S.; Toulza, E.; Lebaron, P.; Lami, R. Diversity of quorum sensing autoinducer synthases in the Global Ocean Sampling Metagenomic Database. Aquat. Microb. Ecol. 2015, 74, 107–119. [Google Scholar] [CrossRef]
- Ziervogel, K.; Arnosti, C. Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate-free seawater from the northeastern Gulf of Mexico. Environ. Microbiol. 2008, 10, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; De Nys, R.; Manefield, M.; Gram, L.; Maiximilien, L.E.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic singalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Teplitski, M.; Robinson, J.B.; Bauer, W.D. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol. Plant Microbe Interact. 2003, 16, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Pujol, I.; Ballester, F.; Joven, J.; Simó, J.M. Paraoxonases as potential antibiofilm agents: Their relationship with quorum-sensing signals in Gram-negative bacteria. Antimicrob. Agents Chemother. 2011, 55, 1325–1331. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Zhang, L.H. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 2005, 43, 101–109. [Google Scholar] [PubMed]
- Romero, M.; Mayer, C.; Muras, A.; Otero, A. Silencing bacterial communication through enzymatic quorum sensing inhibition. In Quorum Sensing vs Quorum Quenching: A Battle with No End in Sight; Kalia, V.C., Ed.; Springer: Delhi, India, 2015; pp. 219–236. [Google Scholar]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Kjelleberg, S.; McDougald, D.; Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibition. In Chemical Communication among Bacteria; Winans, S.C., Bassler, B.L., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 393–416. [Google Scholar]
- Dong, Y.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef]
- Dong, Y.H.; Gusti, A.R.; Zhang, Q.; Xu, J.L.; Zhang, L.H. Identification of quorum-quenching N-acyl homoserine lactoneses from Bacillus species. Appl. Environ. Microbiol. 2002, 68, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo-Picard, C.; Faure, D.; Penot, I.; Dessaux, Y. Diversity of N-acyl homoserine lactone-producing and–degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 2005, 7, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Golberg, K.; Pavlov, V.; Marks, R.S.; Kushmaro, A. Coral-associated bacteria, quorum sensing disrupters, and the regulation of biofouling. Biofouling 2013, 29, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdorf, I.; Oliveiro, G.; Constantino, V.; Morgenstern, D.; Steindler, L. In search of alternative antibiotics drugs: Quorum-quenching activity in sponges and their bacterial isolates. Front. Microbiol. 2016, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Martín-Cuadrado, A.M.; Roca-Rivada, A.; Cabello, A.M.; Otero, A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 2011, 75, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Martín-Cuadrado, A.B.; Otero, A. Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 2012, 78, 6345–6348. [Google Scholar] [CrossRef] [PubMed]
- Linthorne, J.S.; Chang, B.J.; Flematti, G.R.; Ghisalberti, E.L.; Sutton, D.C. A direct pre-screen for marine bacteria producing compounds inhibiting quorum sensing reveals diverse planktonic bacteria that are bioactive. Mar. Biotechol. 2014, 17, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Romero, M.; Prado, S.; Dubert, J.; Tahrioui, A.; Otero, A.; Llamas, I. N-acylhomoserine lactone-degrading bacteria isolated from hatchery bivalve cultures. Microbiol. Res. 2013, 168, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Rubio-Portillo, E.; Antón, J.; Ramos-Esplá, A.A.; Quesada, E.; Llamas, I. Selection of the N-acylhomoserine lactone-degrading bacterium Alteromonas stellopolaris PQQ-42 and of its potential for biocontrol in aquaculture. Front. Microbiol. 2016, 7, 646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skindersoe, M.E.; Ettinger-Esptein, P.; Rasmussen, T.B.; Bjarnsholt, T.; De Nys, R.; Givskov, M. Quorum sensing antagonism from marine organisms. Mar. Biotechnol. 2008, 10, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Hmelo, L.R.; Van Mooy, B.A.S. Kinetic constrainst on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 2009, 54, 127–133. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Pinnow, N.; Schmitz, R.A. Novel reporter for identification of interference with acyl homoserine lactone and autoinducer-2 quorum sensing. Appl. Environ. Microbiol. 2015, 81, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, M.; Haro-Moreno, J.M.; Gonzalez-Serrano, R.; Parras-Moltó, M.; Rodriguez-Valera, F. Genome diversity of marine phages recovered from Mediterranean metagenomes: Size matters. PLoS Genet. 2017, 13, e1007018. [Google Scholar] [CrossRef] [PubMed]
- Haro-Moreno, J.M.; López-Pérez, M.; de la Torre, J.; Picazo, A.; Camacho, A.; Rodriguez-Valera, F. Fine Stratification of Microbial Communities through A Metagenomic Profile of the Photic Zone. ISME J. 2017. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into new bacterial taxonomiy. Appl. Environ. Microbiol. 2007, 73, 5261–5627. [Google Scholar] [CrossRef] [PubMed]
- Twigg, M.S.; Tait, K.; Williams, P.; Atkinson, S.; Cámara, M. Interference with the germination and growth of Ulva zoospores by quorum-sensing molecules from Ulva-associated epiphytic bacteria. Environ. Microbiol. 2014, 16, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W.; Chun, J. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef] [PubMed]
- Chilton, M.D.; Currier, T.C.; Farrand, S.K.; Bendich, A.J.; Gordon, M.P.; Nester, E.W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detectedin crown gall tumors. Proc. Natl. Acad. Sci. USA 1974, 71, 3672–3676. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.D.; Ping, G.; Daly, S.L.; Cha, C.; Cronan, J.E., Jr.; Rinehart, K.L.; Farrand, S.K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 1997, 94, 6036–6041. [Google Scholar] [CrossRef] [PubMed]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; et al. N-acylhomoserine lactonase undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis NS Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Kato, M.; Fukamachi, K.; Kato, N.; Ikeda, T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 2008, 279, 124–130. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Cámara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Romero, M.; Muras, A.; Otero, A. Aii20J, a wide spectrum thermo-stable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 9523–9539. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Muras, A.; Mayer, C.; Buján, N.; Magariños, B.; Otero, A. In vitro quenching of fish pathogen Edwardsiella tarda AHL production using the marine bacterium Tenacibaculum sp. strain 20J cell extracts. Dis. Aquat. Organ. 2014, 108, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Martín-Cuadrado, A.B.; Rodriguez-Valera, F.; Moreira, D.; Alba, J.C.; Ivars-Martinez, E.; Henn, M.R.; Talla, E.; López-García, P. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008, 2, 865–886. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and traslation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.T.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E. Structural RAN Homology Search and Alignment Using Covariance Models. Ph.D. Thesis, Arts and Sciences of Washington University, Saint Louis, MO, USA, December 2009. [Google Scholar] [CrossRef]
- Huang, Y.; Gilna, P.; Li, W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 2009, 25, 1338–1340. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankaaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.Y.; Feodorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 28, 22–28. [Google Scholar] [CrossRef]
- Haft, D.H.; Loftus, B.J.; Richardson, D.L.; Yang, F.; Eisen, J.A.; Paulsen, I.T.; White, O. TIGRFAMs: A protein family resources for the functional identification of proteins. Nucleic Acid Res. 2001, 29, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Multiple alignment using hidden Markov models. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Cambridge, UK, 16–19 July 1995; Volume 3, pp. 114–120. [Google Scholar]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.; Parris, D.J.; DeLong, E.F.; Stewart, F.J. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014, 8, 187–211. [Google Scholar] [CrossRef] [PubMed]
- Cases, I.; de Lorenzo, V.; Ouzounis, C.A. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003, 11, 248–253. [Google Scholar] [CrossRef]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminiscent system. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [PubMed]
- Amann, R.I.; Ludwig, W.; Scheleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 95, 143–169. [Google Scholar]
- Acinas, S.G.; Antón, J.; Rodríguez-Valera, F. Diversity of Free-Living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1999, 65, 514–522. [Google Scholar] [PubMed]
- Diggle, S.P.; Griffin, A.S.; Campbell, G.S.; West, S.A. Cooperation and conflict in quorum sensing bacterial populations. Nature 2007, 450, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Diggle, S.P.; Heeb, S.; Cámara, M.; Otero, A. Quorum quenching activiy in Anabaena sp. PCC7120: Identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 2008, 280, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Pinardi, N.; Masetti, E. Variability of the large scale general circulation of the Mediterranean Sea from observations and modelling: A review. Palaerogeogr. Palaeoclimatol. Palaeoecol. 2000, 158, 153–173. [Google Scholar] [CrossRef]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Ong, S.L.; Leadbetter, J.R.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Norman, R.S.; Visscher, P.T. Quorum sensing in natural environments: Emerging views from microbial mats. Trends Microbiol. 2010, 18, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.N.; Chen, C.J.; Liao, C.T.; Lee, C.Y. A probable aculeacin A Acylase from Ralstonia solanacearum GMI1000 is N-acyl homoserine lactone acylase with quorum-quenching activity. BMC Microbiol. 2009, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Someya, N.; Ikeda, T. Novel N-acylhomoserine lactone-degrading bacteria from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 2009, 73, 2124–2127. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Z.; Morohoshi, T.; Ikenoya, M.; Someya, N.; Ikeda, T. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 2010, 76, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Last, D.; Krüger, G.H.; Dörr, M.; Bornscheuer, U.T. Fast, continuous, and high-throughput (bio)chemical activity assay for N-acyl-l-homoserine lactone quorum-quenching enzymes. Appl. Environ. Microbiol. 2016, 82, 4145–4154. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, K.; Fernandes, M.M.; Francesko, A.; Mendoza, E.; Guezquez, J.; Burnet, M.; Tzanov, T. Quorum-Quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces 2015, 7, 27066–27077. [Google Scholar] [CrossRef] [PubMed]
- Feugeas, J.P.; Tourret, J.; Launay, A.; Bouvet, O.; Hoede, C.; Denamur, E.; Tenaillon, O. Links between transcription, environmental adaptation and gene variability in Escherichia coli: Correlations between gene expression and gene variability reflect growth efficiencies. Mol. Biol. Evol. 2016, 33, 2515–2529. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kang, H.O.; Jang, H.S.; Lee, J.K.; Koo, B.T.; Yum, D.Y. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 2005, 71, 2636–2641. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, R.; Varshney, N.K.; Panigrahi, P.; Suresh, C.G.; Prabhune, A. A new role for penicillin acylases: Degradation of acyl homoserine lactone quorum sensing signals by Kluyvera citrophila penicillin G acylase. Enzym. Microb. Technol. 2014, 56, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Case, R.; Labbate, M.; Kjelleberg, S. AHL-driven quorum-sensing circuits: Their frequency and function among the Proteobacteria. ISME J. 2008, 2, 345–349. [Google Scholar] [CrossRef] [PubMed]


| Live Cell | Cell Extract | MAC C6-HSL (µg Protein/mL Cell Extract) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Closest Cultivated Bacteria | % ID at 16S rRNA Gene Locus | C6-HSL | OC6-HSL | C12-HSL | OC12-HSL | C6-HSL | C12-HSL | ||
| 90 m | 1F1 | Planomicrobium chinense | 99.93 | + | + | + | + | + | + | 104.7 |
| 2E12 | Sphingopyxis alaskensis | 99.93 | + | + | + | + | + | + | 18.67 | |
| 2G12 | Erythrobacter citreus | 99.04 | + | + | + | + | + | + | 1918 | |
| 3A3 | Microbacterium schleiferi | 99.71 | + | + | + | + | - | + | nd | |
| 2000 m | 2F1 | Ralstonia pickettii | 95.79 | + | + | ± | + | + | + | 17.6 |
| 3G7 | Erythrobacter flavus | 99.34 | + | + | + | + | + | + | 786 | |
| 4B4 | Erythrobacter flavus | 99.34 | + | + | + | + | + | + | 216 | |
| 4B7 | Pantoea sp. | 96.01 | + | - | + | - | - | - | nd | |
| 4B9 | Erythrobacter flavus | 99.71 | + | + | + | + | + | + | 961 | |
| 4B10 | Erythrobacter flavus | 99.49 | + | + | + | + | + | + | 223.9 | |
| 4B12 | Erythrobacter flavus | 99.49 | + | + | + | + | + | + | 965 | |
| 4C3 | Citromicrobium sp. | 98.33 | + | + | + | + | - | + | nd | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muras, A.; López-Pérez, M.; Mayer, C.; Parga, A.; Amaro-Blanco, J.; Otero, A. High Prevalence of Quorum-Sensing and Quorum-Quenching Activity among Cultivable Bacteria and Metagenomic Sequences in the Mediterranean Sea. Genes 2018, 9, 100. https://doi.org/10.3390/genes9020100
Muras A, López-Pérez M, Mayer C, Parga A, Amaro-Blanco J, Otero A. High Prevalence of Quorum-Sensing and Quorum-Quenching Activity among Cultivable Bacteria and Metagenomic Sequences in the Mediterranean Sea. Genes. 2018; 9(2):100. https://doi.org/10.3390/genes9020100
Chicago/Turabian StyleMuras, Andrea, Mario López-Pérez, Celia Mayer, Ana Parga, Jaime Amaro-Blanco, and Ana Otero. 2018. "High Prevalence of Quorum-Sensing and Quorum-Quenching Activity among Cultivable Bacteria and Metagenomic Sequences in the Mediterranean Sea" Genes 9, no. 2: 100. https://doi.org/10.3390/genes9020100






